bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-19_CDS_annotation_glimmer3.pl_2_1
Length=326
Score E
Sequences producing significant alignments: (Bits) Value
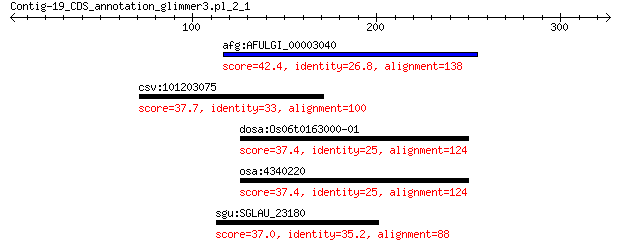
afg:AFULGI_00003040 Lipopolysaccharide biosynthesis protein 42.4 0.15
csv:101203075 putative syntaxin-131-like 37.7 2.4
dosa:Os06t0163000-01 Os06g0163000; Similar to Heat shock prote... 37.4 5.3
osa:4340220 Os06g0163000 37.4 5.3
sgu:SGLAU_23180 large Ala/Glu-rich protein 37.0 6.5
> afg:AFULGI_00003040 Lipopolysaccharide biosynthesis protein
Length=1395
Score = 42.4 bits (98), Expect = 0.15, Method: Composition-based stats.
Identities = 37/139 (27%), Positives = 68/139 (49%), Gaps = 17/139 (12%)
Query 117 DATVNKLNAEAEKLRGDTKDPNVTKDSQRLEFDWNLVKKQREQVQLAVDEINKEFQRANN 176
+A + +LN E E++ G+ KD K+ Q + +L +K +E + V IN E +
Sbjct 338 EAEIERLNDELERISGELKD----KNEQVSQLTRSLEEKTKELNEFKVKSINLEQKIRGL 393
Query 177 EADMQIKRGLYSETLAKIDKLIADKDVSEEMKQNLQKQRDLIESQISATQAQTDFTKAQT 236
E +++ K L + KL + D E+M LQ++ ++S+I+A Q ++
Sbjct 394 EGELKEKESL-------VGKLRRELDEKEKMIGELQQESQKVKSEIAAKDEQI-----KS 441
Query 237 MT-ENMTRDDRIREIKSRI 254
+T EN+ + IKS +
Sbjct 442 LTEENIRLSGELDSIKSSV 460
> csv:101203075 putative syntaxin-131-like
Length=308
Score = 37.7 bits (86), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 33/100 (33%), Positives = 46/100 (46%), Gaps = 5/100 (5%)
Query 71 GNSPGGAGIATDASAPNSQTPMGSSDFNFVTTIAERQRMKNEKAIADATVNKLNAEAEKL 130
G GG I A+AP S G DF E+Q NEK D + KL E+
Sbjct 14 GQPSGGRDIELGANAPTSAGDQGMGDFFKKVQEIEKQ---NEKL--DRLLRKLQDSHEES 68
Query 131 RGDTKDPNVTKDSQRLEFDWNLVKKQREQVQLAVDEINKE 170
+ TK P + QR+E D + V K V+ V+E+++E
Sbjct 69 KAVTKAPAMKAIKQRMEKDVDEVGKVARYVKTKVEELDRE 108
> dosa:Os06t0163000-01 Os06g0163000; Similar to Heat shock protein
STI (Stress inducible protein) (GmSTI).
Length=805
Score = 37.4 bits (85), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 31/132 (23%), Positives = 70/132 (53%), Gaps = 8/132 (6%)
Query 126 EAEKLRGDTKDPNVTKDSQRLEFDWNL--VKKQREQVQLAVDEINKEFQRANNE-----A 178
E+E+L +D +++ QR E + +L V+++ EQ+++ DE+ E QR N +
Sbjct 322 ESERLARTLEDLYLSQVQQRKETEESLSRVQQEFEQLKIQQDEVTVELQRVNEQNENLLG 381
Query 179 DMQIKRGLYSETLAKIDKLIADKD-VSEEMKQNLQKQRDLIESQISATQAQTDFTKAQTM 237
+ R + L++ D+L+ ++D E+++ QK+ ++ ++A + ++ ++
Sbjct 382 QLSDSREHFEWLLSEHDQLLRERDNAVREVEELRQKRGQMLSVLVTAMHCEFSSSEVESA 441
Query 238 TENMTRDDRIRE 249
TEN + +I E
Sbjct 442 TENFSNSLKIGE 453
> osa:4340220 Os06g0163000
Length=805
Score = 37.4 bits (85), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 31/132 (23%), Positives = 70/132 (53%), Gaps = 8/132 (6%)
Query 126 EAEKLRGDTKDPNVTKDSQRLEFDWNL--VKKQREQVQLAVDEINKEFQRANNE-----A 178
E+E+L +D +++ QR E + +L V+++ EQ+++ DE+ E QR N +
Sbjct 322 ESERLARTLEDLYLSQVQQRKETEESLSRVQQEFEQLKIQQDEVTVELQRVNEQNENLLG 381
Query 179 DMQIKRGLYSETLAKIDKLIADKD-VSEEMKQNLQKQRDLIESQISATQAQTDFTKAQTM 237
+ R + L++ D+L+ ++D E+++ QK+ ++ ++A + ++ ++
Sbjct 382 QLSDSREHFEWLLSEHDQLLRERDNAVREVEELRQKRGQMLSVLVTAMHCEFSSSEVESA 441
Query 238 TENMTRDDRIRE 249
TEN + +I E
Sbjct 442 TENFSNSLKIGE 453
> sgu:SGLAU_23180 large Ala/Glu-rich protein
Length=1326
Score = 37.0 bits (84), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 31/88 (35%), Positives = 48/88 (55%), Gaps = 5/88 (6%)
Query 113 KAIADATVNKLNAEAEKLRGDTKDPNVTKDSQRLEFDWNLVKKQREQVQLAVDEINKEFQ 172
+A A TV A AE++R +++ VT S+ E L RE+ + +DE KE
Sbjct 1008 RAEAAETVGSAQAHAERVRAESE--RVT--SEAAEEAERLTTAAREEAERTLDEARKEAN 1063
Query 173 RANNEADMQIKRGLYSETLAKIDKLIAD 200
+ +EA Q+ + L SET A+ DKL+A+
Sbjct 1064 KRRSEAAEQVDK-LISETAAEADKLLAE 1090
Lambda K H a alpha
0.311 0.125 0.335 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 581865132318