bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-1_CDS_annotation_glimmer3.pl_2_5
Length=176
Score E
Sequences producing significant alignments: (Bits) Value
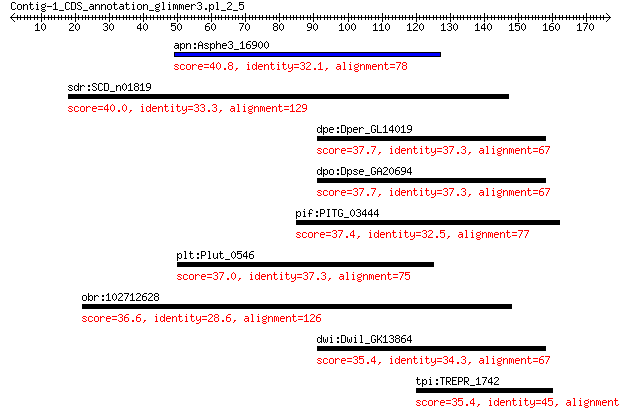
apn:Asphe3_16900 beta-glucosidase/6-phospho-beta-glucosidase/b... 40.8 0.088
sdr:SCD_n01819 TRAP dicarboxylate transporter subunit DctP 40.0 0.13
dpe:Dper_GL14019 GL14019 gene product from transcript GL14019-RA 37.7 0.94
dpo:Dpse_GA20694 GA20694 gene product from transcript GA20694-RA 37.7 0.98
pif:PITG_03444 dynein heavy chain 37.4 1.7
plt:Plut_0546 glycosyl transferase 37.0 1.7
obr:102712628 inactive poly [ADP-ribose] polymerase RCD1-like 36.6 2.5
dwi:Dwil_GK13864 GK13864 gene product from transcript GK13864-RA 35.4 5.2
tpi:TREPR_1742 2,3-cyclic-nucleotide 2'phosphodiesterase (EC:3... 35.4 6.4
> apn:Asphe3_16900 beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase
Length=428
Score = 40.8 bits (94), Expect = 0.088, Method: Compositional matrix adjust.
Identities = 25/81 (31%), Positives = 37/81 (46%), Gaps = 3/81 (4%)
Query 49 ETIGEGPDKYYRYRSDISMLLHAKDTANKIGIEGLRYLSESRRTKTSAI---QSQLDQMD 105
E G+ D Y+RYR DI+++ T+ + IE R E R T+ + + L+
Sbjct 44 EPSGDAVDHYHRYREDIALIAGLGFTSYRFSIEWARVEPEEGRFSTAELDHYRRMLEACR 103
Query 106 DQLLLDTVKSRHLQSPSEILA 126
D L V H SP +LA
Sbjct 104 DHGLTPVVTFHHFTSPRWLLA 124
> sdr:SCD_n01819 TRAP dicarboxylate transporter subunit DctP
Length=337
Score = 40.0 bits (92), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 43/148 (29%), Positives = 67/148 (45%), Gaps = 22/148 (15%)
Query 18 VDPSISAVGFNPAVRE-KEIAVEI---------DPVSKFCIETIGEGPDKYYRYRSDISM 67
+D ++ A+G NP V E+ + +PVS F + + E + Y S +
Sbjct 175 LDATMKALGANPQVMAFSEVYTGLQQGVVDGTENPVSNFYTQKMNEV--QKYMTISQHNY 232
Query 68 LLHAKDTANKIGIEGL----RYLSESRRTKTSAIQSQLDQMDDQLLLDTVKSR-----HL 118
L +A NK EGL R ES +T+A + + Q ++ L VK+ H
Sbjct 233 LGYAV-VVNKKFWEGLPVDVRTALESAMKETTAFERDIAQQENDEALAKVKAAKTTDIHY 291
Query 119 QSPSEILAWSESLIQVAHELETRVGAEI 146
QSP E L W ++L+ V E E +G +I
Sbjct 292 QSPKERLDWYKALLPVHKEFEDVIGKDI 319
> dpe:Dper_GL14019 GL14019 gene product from transcript GL14019-RA
Length=765
Score = 37.7 bits (86), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 36/67 (54%), Gaps = 7/67 (10%)
Query 91 RTKTSAIQSQLDQMDDQLLLDTVKSRHLQSPSEILAWSESLIQVAHELETRVGAEIRKKY 150
R +S++ +M + + LDT+K+ SP L W+ +L H LE R G E+ KKY
Sbjct 22 RPPVPGEESRVKKMTEGVALDTLKN----SPPSYLNWARTL---NHLLEDRDGVELFKKY 74
Query 151 DEEIAAA 157
EE A A
Sbjct 75 VEEEAPA 81
> dpo:Dpse_GA20694 GA20694 gene product from transcript GA20694-RA
Length=762
Score = 37.7 bits (86), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 36/67 (54%), Gaps = 7/67 (10%)
Query 91 RTKTSAIQSQLDQMDDQLLLDTVKSRHLQSPSEILAWSESLIQVAHELETRVGAEIRKKY 150
R +S++ +M + + LDT+K+ SP L W+ +L H LE R G E+ KKY
Sbjct 22 RPPVPGEESRVKKMTEGVALDTLKN----SPPSYLNWARTL---NHLLEDRDGVELFKKY 74
Query 151 DEEIAAA 157
EE A A
Sbjct 75 VEEEAPA 81
> pif:PITG_03444 dynein heavy chain
Length=3965
Score = 37.4 bits (85), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 46/80 (58%), Gaps = 4/80 (5%)
Query 85 YLSESRRTKTSAIQSQLDQMDDQLL-LDTVKSRHLQS--PSEILAWSESLIQVAHELETR 141
Y +E R +T+A++ +D Q L L +V RH+ + +++ W+ L+Q + R
Sbjct 799 YRAEMERVQTTALRLAIDNRHSQHLGLFSVDCRHVNTRLHTDLARWTIRLLQAFEQRTGR 858
Query 142 VGAEIRKKYDEEIAAAAAAS 161
+ AE+R++Y +EIAA A +
Sbjct 859 MNAELRQQY-KEIAARLAKT 877
> plt:Plut_0546 glycosyl transferase
Length=348
Score = 37.0 bits (84), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 43/89 (48%), Gaps = 14/89 (16%)
Query 50 TIGEG-PDK-YYRYRSDISMLLHAKDTANKIGIEGLRYLSE------------SRRTKTS 95
++GEG P+K Y+ +R+++ MLL + T I LRYL E R K +
Sbjct 243 SLGEGSPEKIYFNHRNNMLMLLKNRSTGGLFRILPLRYLLEFAAVLYYLPHAPEGRRKAA 302
Query 96 AIQSQLDQMDDQLLLDTVKSRHLQSPSEI 124
A+ + QL L K RH+QS ++
Sbjct 303 AVFRAMRDAFLQLPLTLRKRRHVQSSRKV 331
> obr:102712628 inactive poly [ADP-ribose] polymerase RCD1-like
Length=871
Score = 36.6 bits (83), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 36/131 (27%), Positives = 65/131 (50%), Gaps = 15/131 (11%)
Query 22 ISAVGFNPAVREKEIAVEIDPVSKFCIETIGEGP-DKYYRYRSDISMLLHAKDTANKIGI 80
+ + F+P+ KE+AV+I + ET EGP + +R +M+ +D + +
Sbjct 403 LKGLWFHPS--PKEVAVDISTLVPIMSET-AEGPTSPWISFRVLFAMI---QDNISSVAR 456
Query 81 EGLRY----LSESRRTKTSAIQSQLDQMDDQLLLDTVKSRHLQSPSEILAWSESLIQVAH 136
E L + L E++ T+ ++ + + ++LLL+T+K H PS W +S ++VA
Sbjct 457 ELLFHHYEELKENKITREEMVKQMIILVGEKLLLETLKRLHY-CPS---LWYKSSVEVAS 512
Query 137 ELETRVGAEIR 147
R AE R
Sbjct 513 SDPARTAAEDR 523
> dwi:Dwil_GK13864 GK13864 gene product from transcript GK13864-RA
Length=748
Score = 35.4 bits (80), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 35/67 (52%), Gaps = 7/67 (10%)
Query 91 RTKTSAIQSQLDQMDDQLLLDTVKSRHLQSPSEILAWSESLIQVAHELETRVGAEIRKKY 150
R +S++ +M + +++D K+ SP L W+ +L H LE R G E+ KKY
Sbjct 20 RPPVPGEESRVKKMTEGVIIDNSKN----SPPSYLNWARTL---NHLLEDRDGVELFKKY 72
Query 151 DEEIAAA 157
EE A A
Sbjct 73 VEEEAPA 79
> tpi:TREPR_1742 2,3-cyclic-nucleotide 2'phosphodiesterase (EC:3.1.4.16)
Length=656
Score = 35.4 bits (80), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 25/40 (63%), Gaps = 2/40 (5%)
Query 120 SPSEILAWSESLIQVAHELETRVGAEIRKKYDEEIAAAAA 159
+P +I+ W SL+Q ++ETR G E K+Y EI AA A
Sbjct 195 TPPQIITWDGSLLQ--GKVETRDGYETAKRYIPEIKAAGA 232
Lambda K H a alpha
0.312 0.128 0.349 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 150452743398