bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-25_CDS_annotation_glimmer3.pl_2_1
Length=174
Score E
Sequences producing significant alignments: (Bits) Value
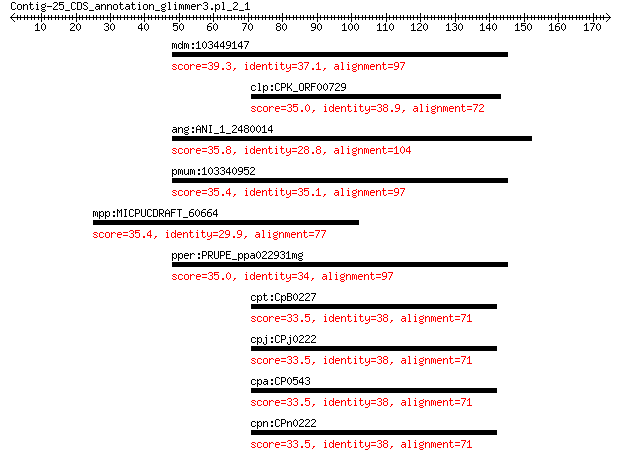
mdm:103449147 ARF guanine-nucleotide exchange factor GNL2-like 39.3 0.26
clp:CPK_ORF00729 hypothetical protein 35.0 2.5
ang:ANI_1_2480014 An01g04470; PHD finger and BAH domain protei... 35.8 4.6
pmum:103340952 ARF guanine-nucleotide exchange factor GNL2 35.4 6.5
mpp:MICPUCDRAFT_60664 ATP-binding cassette superfamily 35.4 6.9
pper:PRUPE_ppa022931mg hypothetical protein 35.0 8.9
cpt:CpB0227 hypothetical protein 33.5 9.5
cpj:CPj0222 hypothetical protein 33.5 9.5
cpa:CP0543 hypothetical protein 33.5 9.5
cpn:CPn0222 hypothetical protein 33.5 9.5
> mdm:103449147 ARF guanine-nucleotide exchange factor GNL2-like
Length=1384
Score = 39.3 bits (90), Expect = 0.26, Method: Composition-based stats.
Identities = 36/125 (29%), Positives = 53/125 (42%), Gaps = 42/125 (34%)
Query 48 SPLQRFKDEYFEELVWMLPELAESLKKKNNTDANGAFPQFKGLLKYINIRDYQLFSK--- 104
SPL + + FE LV M+ +A+S+ K+N+T +G +P I IR+Y F +
Sbjct 433 SPLTTLQIQAFEGLVIMIHNIADSIDKENDTSPSGPYP--------IEIREYTPFWEDKP 484
Query 105 -----------RLRKYLSKKI--------------GKYEKIHSYVVSEYSPKTLRPHFHI 139
RLRK +KI +Y K+ + V PKTL
Sbjct 485 KDDSEAWVQFVRLRKAQKRKILIAGHHFNCDEKKGMEYLKLSNLVSDPPDPKTLA----- 539
Query 140 LFFFR 144
+FFR
Sbjct 540 -YFFR 543
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 35.0 bits (79), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 28/76 (37%), Positives = 36/76 (47%), Gaps = 13/76 (17%)
Query 71 SLKKKNN----TDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVS 126
SL +KN T + PQ+ L+K QLF KRLR +S KI +
Sbjct 47 SLYEKNCFLTLTYDDKHLPQYGSLVKL----HLQLFLKRLRDRISP-----HKIRYFGCG 97
Query 127 EYSPKTLRPHFHILFF 142
EY K RPH+H+L F
Sbjct 98 EYGTKLQRPHYHLLIF 113
> ang:ANI_1_2480014 An01g04470; PHD finger and BAH domain protein
(Snt2)
Length=1673
Score = 35.8 bits (81), Expect = 4.6, Method: Composition-based stats.
Identities = 30/121 (25%), Positives = 55/121 (45%), Gaps = 24/121 (20%)
Query 48 SPLQR--FKDEYFEELVWMLPELAE------------SLKKKNNTDANGAFPQFKGLLKY 93
SP +R F D+Y E + P+L E L N+ DA+ A + K + KY
Sbjct 647 SPAEREKFVDDYMERAKALAPQLGEEKYATNFLDKALELLYSNSFDADAALAKLKTMNKY 706
Query 94 INIRDYQLFSKRLRKY---LSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFFFRLGRNRP 150
++++ L + L+ + +SK ++ I YV + PH+ I+ F+ + + P
Sbjct 707 KDLKEPHLRPEELKLFEQAVSKFGSEWRNITKYVGTV-------PHYQIVRFYYMWKKTP 759
Query 151 K 151
+
Sbjct 760 R 760
> pmum:103340952 ARF guanine-nucleotide exchange factor GNL2
Length=1387
Score = 35.4 bits (80), Expect = 6.5, Method: Composition-based stats.
Identities = 34/125 (27%), Positives = 54/125 (43%), Gaps = 42/125 (34%)
Query 48 SPLQRFKDEYFEELVWMLPELAESLKKKNNTDANGAFPQFKGLLKYINIRDYQLFSK--- 104
+PL + + FE LV M+ +AES+ ++++T +G +P I I +Y F +
Sbjct 434 NPLTTIQIQAFEGLVIMIHNIAESIDREHDTSPSGPYP--------IEITEYAPFWEDKP 485
Query 105 -----------RLRKYLSKKI-------GKYE-------KIHSYVVSEYSPKTLRPHFHI 139
R+RK +KI +YE K+++ V PKTL
Sbjct 486 KDDSEAWVQFVRVRKAQKRKILIAGHHFNRYENKGMEYLKLYNLVSDPPDPKTLA----- 540
Query 140 LFFFR 144
FFFR
Sbjct 541 -FFFR 544
> mpp:MICPUCDRAFT_60664 ATP-binding cassette superfamily
Length=2018
Score = 35.4 bits (80), Expect = 6.9, Method: Composition-based stats.
Identities = 23/77 (30%), Positives = 34/77 (44%), Gaps = 4/77 (5%)
Query 25 LSLRYPNFGSKFRPYILRSILRKSPLQRFKDEYFEELVWMLPELAESLKKKNNTDANGAF 84
L LR PN R + R + R+ P R + + + + LP +A L + GAF
Sbjct 1924 LDLRVPNVADVAR--VRRHVEREIPGARETEAHASRVRYRLPSMA--LARGGKASVAGAF 1979
Query 85 PQFKGLLKYINIRDYQL 101
+G + I DYQL
Sbjct 1980 ASLEGTKTRLGIEDYQL 1996
> pper:PRUPE_ppa022931mg hypothetical protein
Length=1384
Score = 35.0 bits (79), Expect = 8.9, Method: Composition-based stats.
Identities = 33/125 (26%), Positives = 53/125 (42%), Gaps = 42/125 (34%)
Query 48 SPLQRFKDEYFEELVWMLPELAESLKKKNNTDANGAFPQFKGLLKYINIRDYQLFSK--- 104
+PL + + FE LV M+ +AES+ ++++T +G +P I I +Y F +
Sbjct 434 NPLTTIQIQAFEGLVIMIHNIAESIDREHDTSPSGPYP--------IEITEYAPFWEDKP 485
Query 105 -----------RLRKYLSKKI--------------GKYEKIHSYVVSEYSPKTLRPHFHI 139
R+RK +KI +Y K+++ V PKTL
Sbjct 486 KDDSEAWVQFVRVRKAQKRKILIAGHHFNRDEKKGMEYLKLYNLVSDPPDPKTLA----- 540
Query 140 LFFFR 144
FFFR
Sbjct 541 -FFFR 544
> cpt:CpB0227 hypothetical protein
Length=113
Score = 33.5 bits (75), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 35/75 (47%), Gaps = 13/75 (17%)
Query 71 SLKKKNN----TDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVS 126
SL +KN T + PQ+ L+K QLF KRLRK +S KI +
Sbjct 47 SLYEKNCFLTLTYDDKHLPQYGSLVKL----HLQLFLKRLRKMISP-----HKIRYFECG 97
Query 127 EYSPKTLRPHFHILF 141
Y K RPH+H+L
Sbjct 98 AYGTKLQRPHYHLLL 112
> cpj:CPj0222 hypothetical protein
Length=113
Score = 33.5 bits (75), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 35/75 (47%), Gaps = 13/75 (17%)
Query 71 SLKKKNN----TDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVS 126
SL +KN T + PQ+ L+K QLF KRLRK +S KI +
Sbjct 47 SLYEKNCFLTLTYDDKHLPQYGSLVKL----HLQLFLKRLRKMISP-----HKIRYFECG 97
Query 127 EYSPKTLRPHFHILF 141
Y K RPH+H+L
Sbjct 98 AYGTKLQRPHYHLLL 112
> cpa:CP0543 hypothetical protein
Length=113
Score = 33.5 bits (75), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 35/75 (47%), Gaps = 13/75 (17%)
Query 71 SLKKKNN----TDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVS 126
SL +KN T + PQ+ L+K QLF KRLRK +S KI +
Sbjct 47 SLYEKNCFLTLTYDDKHLPQYGSLVKL----HLQLFLKRLRKMISP-----HKIRYFECG 97
Query 127 EYSPKTLRPHFHILF 141
Y K RPH+H+L
Sbjct 98 AYGTKLQRPHYHLLL 112
> cpn:CPn0222 hypothetical protein
Length=113
Score = 33.5 bits (75), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 35/75 (47%), Gaps = 13/75 (17%)
Query 71 SLKKKNN----TDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVS 126
SL +KN T + PQ+ L+K QLF KRLRK +S KI +
Sbjct 47 SLYEKNCFLTLTYDDKHLPQYGSLVKL----HLQLFLKRLRKMISP-----HKIRYFECG 97
Query 127 EYSPKTLRPHFHILF 141
Y K RPH+H+L
Sbjct 98 AYGTKLQRPHYHLLL 112
Lambda K H a alpha
0.324 0.139 0.425 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 143911319772