bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-25_CDS_annotation_glimmer3.pl_2_5
Length=242
Score E
Sequences producing significant alignments: (Bits) Value
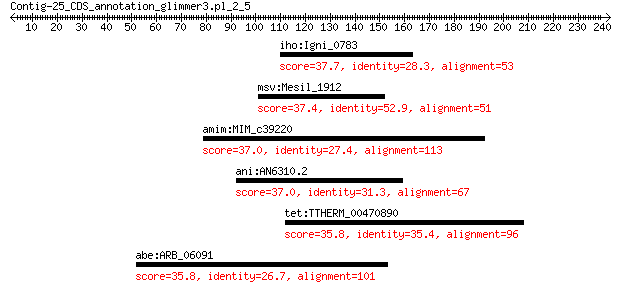
iho:Igni_0783 hypothetical protein 37.7 1.4
msv:Mesil_1912 hypothetical protein 37.4 2.4
amim:MIM_c39220 putative succinate-semialdehyde dehydrogenase 37.0 2.9
ani:AN6310.2 hypothetical protein 37.0 4.6
tet:TTHERM_00470890 Protein kinase domain containing protein 35.8 8.5
abe:ARB_06091 PAP/25A associated domain family protein 35.8 8.7
> iho:Igni_0783 hypothetical protein
Length=294
Score = 37.7 bits (86), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 32/53 (60%), Gaps = 0/53 (0%)
Query 110 GSNMPCLSADQISQSLDQLKSKQNYFGFDRADLAYKLLQYLRYGNVRTGVGSN 162
GS +P + AD++ ++ D LK + + +L YK+L++ ++ + R G+G +
Sbjct 69 GSEVPLVCADEVVEAPDPLKRFEEVLSAPKDELEYKVLKFSKFLSGRVGLGGS 121
> msv:Mesil_1912 hypothetical protein
Length=511
Score = 37.4 bits (85), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 33/64 (52%), Gaps = 15/64 (23%)
Query 101 SSFDGSVLLGSNMPCLSADQISQSLDQLKSKQNY-------------FGFDRADLAYKLL 147
SSFDGS GSN L+ D+ISQ L + +K Y + FDRADLA +L
Sbjct 301 SSFDGSG--GSNAMALTLDKISQRLTRSDAKAVYSTLRRDLVVPFCHYNFDRADLAPRLE 358
Query 148 QYLR 151
LR
Sbjct 359 PILR 362
> amim:MIM_c39220 putative succinate-semialdehyde dehydrogenase
Length=477
Score = 37.0 bits (84), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 31/116 (27%), Positives = 49/116 (42%), Gaps = 15/116 (13%)
Query 79 LHLLWRNAPEVIS-QIQQNVQHASSFDGSVLLGSNMPCLSADQISQSLDQLKSKQNYFGF 137
L+++W + P+V I+ + SF GSV +G + L+ + + +L F
Sbjct 202 LNIVWGDPPKVSDYLIRSPIVRKVSFTGSVPVGKQLAALAGAHMKRITMELGGHSPVLVF 261
Query 138 DRADL--AYKLLQYLRYGNVRTGVGSNGARNYGTSIDVKDGTYNQNRAYNHALSIF 191
D AD+ A K+L + RN G Y Q +AY+ LSIF
Sbjct 262 DDADIPKAAKMLAKFKI------------RNAGQVCISPTRFYVQEKAYDQFLSIF 305
> ani:AN6310.2 hypothetical protein
Length=4917
Score = 37.0 bits (84), Expect = 4.6, Method: Composition-based stats.
Identities = 21/67 (31%), Positives = 35/67 (52%), Gaps = 2/67 (3%)
Query 92 QIQQNVQHASSFDGSVLLGSNMPCLSADQISQSLDQLKSKQNYFGFDRADLAYKLLQYLR 151
+++Q+++H V S + + D + SLDQLK+ + DR DL + L Q LR
Sbjct 3859 RLRQSIKHLPELPSGVT--SKLHQHTFDDAASSLDQLKTDITKWARDRPDLTFVLDQLLR 3916
Query 152 YGNVRTG 158
+ V+ G
Sbjct 3917 WTKVKMG 3923
> tet:TTHERM_00470890 Protein kinase domain containing protein
Length=994
Score = 35.8 bits (81), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 34/105 (32%), Positives = 47/105 (45%), Gaps = 15/105 (14%)
Query 112 NMPCLSADQIS-----QSLDQLKSKQNYFGFDRADLAYK--LLQYLRYGNVRTGVGSNGA 164
N+P S Q S + ++ L KQN + FD D + K L YL GNV G+ N
Sbjct 556 NLPAQSGQQNSNQSPLEQINLLTQKQNIYLFDYIDCSSKFGLAYYLNNGNV--GILLND- 612
Query 165 RNYGTSIDVKDGTYN--QNRAYNHALSIFPILAYKKFCQDYFRLT 207
G + D T+ Q N+ + +PI CQ+ FRLT
Sbjct 613 ---GNVTQIVDETFEHIQKVQDNYVIQKYPIKQIPPSCQNLFRLT 654
> abe:ARB_06091 PAP/25A associated domain family protein
Length=1179
Score = 35.8 bits (81), Expect = 8.7, Method: Composition-based stats.
Identities = 27/103 (26%), Positives = 45/103 (44%), Gaps = 6/103 (6%)
Query 52 QHFTRTQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEVISQIQQNV-QHASSFDGSVLLG 110
+H+T+ + +N +A Y W ++ L P ++ +Q+ V Q S DGS +
Sbjct 295 KHWTKRRILNDAALGGTLSSYTWICLIINFLQTRIPPIVPSLQKRVAQSEGSTDGSSITS 354
Query 111 SNMPCLSADQISQSLDQLKSKQNYFGFD-RADLAYKLLQYLRY 152
+ S + S D K FG D ++ L L Q+ RY
Sbjct 355 TT----SCNSTYSSFDDDVEKLGGFGDDNKSTLGELLFQFFRY 393
Lambda K H a alpha
0.322 0.136 0.438 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 337638516438