bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-27_CDS_annotation_glimmer3.pl_2_1
Length=197
Score E
Sequences producing significant alignments: (Bits) Value
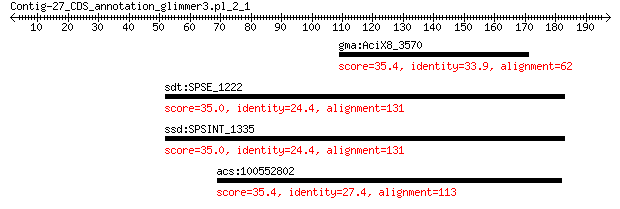
gma:AciX8_3570 xanthine dehydrogenase 35.4 6.6
sdt:SPSE_1222 era; GTP-binding protein Era 35.0 8.5
ssd:SPSINT_1335 GTP-binding protein Era 35.0 8.5
acs:100552802 syne1; spectrin repeat containing, nuclear envel... 35.4 9.0
> gma:AciX8_3570 xanthine dehydrogenase
Length=748
Score = 35.4 bits (80), Expect = 6.6, Method: Composition-based stats.
Identities = 21/62 (34%), Positives = 34/62 (55%), Gaps = 1/62 (2%)
Query 109 DTDTALKQANIRLTDAQKTQVEHYTNSIDIHRDAAVFKLQQDQKYDDAQRIVTVANQATQ 168
DT TALK A +++ + T VE + N I++H AV+ Q+ Y+ +Q +V + Q
Sbjct 162 DTATALKSAKVKVDEVYTTPVETH-NPIELHASVAVYDGQKFTLYETSQAVVNHRDVLAQ 220
Query 169 SL 170
L
Sbjct 221 ML 222
> sdt:SPSE_1222 era; GTP-binding protein Era
Length=298
Score = 35.0 bits (79), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 32/132 (24%), Positives = 57/132 (43%), Gaps = 14/132 (11%)
Query 52 DASTIATRTAAILNNRRFDLECEDFARRVRETDAKVNLSEAEAKSILVTMYAKVNNIDTD 111
DA + T I + + D+ +V AK LSE +A +V + ++ D
Sbjct 53 DAQIVFLDTPGIHKPKH---KLGDYMMKV----AKNTLSEIDAVMFMVNVNEEIGRGD-- 103
Query 112 TALKQANIRLTDAQKTQVEHYTNSID-IHRDAAVFKLQQDQKYDDAQRIVTVANQATQSL 170
+ + + KT V N ID +H DA + +++Q Q+Y D I+ ++ ++
Sbjct 104 ----EYIMEMLKTVKTPVFLVLNKIDLVHPDALMPRIEQYQRYMDFAEIIPISALEGHNV 159
Query 171 YHISQVASDWLP 182
H V +LP
Sbjct 160 DHFINVLKSYLP 171
> ssd:SPSINT_1335 GTP-binding protein Era
Length=298
Score = 35.0 bits (79), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 32/132 (24%), Positives = 57/132 (43%), Gaps = 14/132 (11%)
Query 52 DASTIATRTAAILNNRRFDLECEDFARRVRETDAKVNLSEAEAKSILVTMYAKVNNIDTD 111
DA + T I + + D+ +V AK LSE +A +V + ++ D
Sbjct 53 DAQIVFLDTPGIHKPKH---KLGDYMMKV----AKNTLSEIDAVMFMVNVNEEIGRGD-- 103
Query 112 TALKQANIRLTDAQKTQVEHYTNSID-IHRDAAVFKLQQDQKYDDAQRIVTVANQATQSL 170
+ + + KT V N ID +H DA + +++Q Q+Y D I+ ++ ++
Sbjct 104 ----EYIMEMLKTVKTPVFLVLNKIDLVHPDALMPRIEQYQRYMDFAEIIPISALEGHNV 159
Query 171 YHISQVASDWLP 182
H V +LP
Sbjct 160 DHFINVLKSYLP 171
> acs:100552802 syne1; spectrin repeat containing, nuclear envelope
1
Length=8758
Score = 35.4 bits (80), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 31/113 (27%), Positives = 54/113 (48%), Gaps = 16/113 (14%)
Query 69 FDLECEDFARRVRETDAKVNLSEAEAKSILVTMYAKVNNIDTDTALKQANIRLTDAQKTQ 128
DL+ D +R KV + + E +S L M A ++N + + +K + TD + +
Sbjct 4867 IDLKLNDIQEEIR----KVQIQQEEVQSSLRIMRA-LSNKEKEKYMKAKELIPTDLENSL 4921
Query 129 VEHYTNSIDIHRDAAVFKLQQDQKYDDAQRIVTVANQATQSLYHISQVASDWL 181
E + +D AVFK Q+ +++ + + Q YH++Q ASDWL
Sbjct 4922 AE--LSELDEEVQEAVFKRQE--------KLIQL-HSMCQRYYHVTQTASDWL 4963
Lambda K H a alpha
0.313 0.125 0.338 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 210745007395