bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-2_CDS_annotation_glimmer3.pl_2_2
Length=630
Score E
Sequences producing significant alignments: (Bits) Value
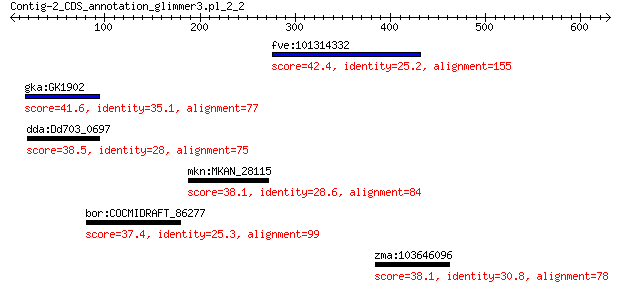
fve:101314332 capsid protein VP1-like 42.4 0.31
gka:GK1902 alpha-D-mannosidase (EC:3.2.1.24) 41.6 0.72
dda:Dd703_0697 glycoside hydrolase family protein 38.5 5.0
mkn:MKAN_28115 membrane protein 38.1 6.8
bor:COCMIDRAFT_86277 hypothetical protein 37.4 8.2
zma:103646096 BEACH domain-containing protein lvsC-like 38.1 9.2
> fve:101314332 capsid protein VP1-like
Length=421
Score = 42.4 bits (98), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 39/155 (25%), Positives = 65/155 (42%), Gaps = 4/155 (3%)
Query 276 FSLRYCNWNKDLFMGVLPNSQFGDIAVIDIEGGLNIPASRISLSSNNRPTIGIKVGAQVS 335
++L D F LP Q G AV + G + P I+ S + +G+ Q +
Sbjct 216 YTLLRRGKRHDYFTSALPWPQKGGTAV-SLPLGTSAP---IAFSGASGSDVGVISTTQGN 271
Query 336 SPNNCSITNSSGNLSTGDILSVGIPAASYKLQSSFNVLALRQAESLQKYREITQSVDTNY 395
N T S +L G A ++ + LRQ+ +QK E T Y
Sbjct 272 LIKNMYSTGSGTSLKIGSATVATGLYADLSAATAATINQLRQSFQIQKLLERDARGGTRY 331
Query 396 RDQIKAHFGVNVPASDSHMAQYIGGIARNLDISEV 430
+ I++HFGV P + +Y+GG + ++I+ +
Sbjct 332 TEIIRSHFGVASPDARLQRPEYLGGGSTPINIAPI 366
> gka:GK1902 alpha-D-mannosidase (EC:3.2.1.24)
Length=1044
Score = 41.6 bits (96), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 40/79 (51%), Gaps = 3/79 (4%)
Query 17 SGFDIGAKN--VFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAYTRIREYFD 74
S +D+ AK + KCG +L V+ D + +DIDI Y + R V+ + I+E
Sbjct 724 SIYDLQAKREVLDDGKCGNVLQVFEDKPLRFDAWDIDIFYQEKKREVEDLQHVSIKEITS 783
Query 75 FYAVPIDLIWKSFDASVIQ 93
YAV I WK D+ + Q
Sbjct 784 LYAV-IHFEWKYMDSIIKQ 801
> dda:Dd703_0697 glycoside hydrolase family protein
Length=1037
Score = 38.5 bits (88), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 35/75 (47%), Gaps = 1/75 (1%)
Query 19 FDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAYTRIREYFDFYAV 78
FD + A CG +L VY D + +DI+I Y + RPV +RE +
Sbjct 720 FDRQHQRDVLADCGNVLTVYEDKPLKYDAWDIEIFYIQKQRPVTELLSATVREQGEL-CC 778
Query 79 PIDLIWKSFDASVIQ 93
I+ +W+ + ++Q
Sbjct 779 SIEFVWRYHHSRIVQ 793
> mkn:MKAN_28115 membrane protein
Length=480
Score = 38.1 bits (87), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 24/84 (29%), Positives = 38/84 (45%), Gaps = 2/84 (2%)
Query 188 RWWNREAPLASDSLIVYSQMYNFNMNVNLFPLATYQKIYQDFFRWSQWEKADPTSYNFDW 247
RWW R A L + L V S N+ V FP T Q + ++ADP S
Sbjct 105 RWWGRGASLMAVPLCVLSAALALNLWVGYFP--TVQAAWNQLTSGPLPDQADPASVAAMA 162
Query 248 YQGSGNLFGGTIDTSLPASSDYWK 271
+G+ G + S+P+++ ++K
Sbjct 163 GKGTRPPRGSVVPVSIPSTASHFK 186
> bor:COCMIDRAFT_86277 hypothetical protein
Length=333
Score = 37.4 bits (85), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 25/106 (24%), Positives = 43/106 (41%), Gaps = 7/106 (7%)
Query 81 DLIWKSFDASVIQMGETAPVQ-------AKDILTALTVSGDLPYCSLSDLGLSCFFASGS 133
D++ ++ A+ + M P + +T+ G L C L L A +
Sbjct 138 DVLEEAIPATAVHMTPMMPTKIHIRHSLESQKRKQITIPGSLSVCDDESLKLGTSLAQEN 197
Query 134 MSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNNMPA 179
P L S + ++ A I G I+GD Y+ + YG I ++ P
Sbjct 198 SIKPDLTSGEEDDTQALIRGAIKGDAEYQTKLLRLYGEISASDGPC 243
> zma:103646096 BEACH domain-containing protein lvsC-like
Length=2865
Score = 38.1 bits (87), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 24/84 (29%), Positives = 48/84 (57%), Gaps = 7/84 (8%)
Query 384 YREITQSVDTNYRDQIKAHFGVNVPASDSHMAQYIGGIARNLDISEVVNNNL----QGDG 439
+ E + TN +D + ++ G VP S++ + + +GGI+ ++ S+ V NN+ +GDG
Sbjct 908 HEEAIEHEATNAKDMLDSNIGSKVPGSENGLLKNLGGISFSI-TSDNVRNNVYNVDKGDG 966
Query 440 EAV--IYGKGVGTGTGSMRYTTGS 461
V I+ G G+G +++ +G+
Sbjct 967 IVVGIIHILGALIGSGHLKFDSGA 990
Lambda K H a alpha
0.319 0.136 0.422 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1472746356532