bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-30_CDS_annotation_glimmer3.pl_2_1
Length=496
Score E
Sequences producing significant alignments: (Bits) Value
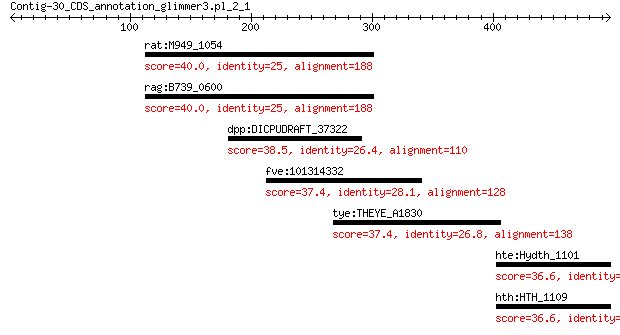
rat:M949_1054 surface antigen (d15) 40.0 1.2
rag:B739_0600 hypothetical protein 40.0 1.2
dpp:DICPUDRAFT_37322 hypothetical protein 38.5 3.5
fve:101314332 capsid protein VP1-like 37.4 8.2
tye:THEYE_A1830 heterodisulfide reductase subunit A/methylviol... 37.4 9.0
hte:Hydth_1101 uroporphyrinogen III synthase HEM4 36.6 9.2
hth:HTH_1109 hemD2; uroporphyrinogen-III synthase 36.6 9.2
> rat:M949_1054 surface antigen (d15)
Length=510
Score = 40.0 bits (92), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 47/204 (23%), Positives = 88/204 (43%), Gaps = 30/204 (15%)
Query 113 LEFLDAFFESQFYPSAVASTNNTFNRGNLF------YQIICSDLDSTGSTGDGFPVKNIY 166
++FLD+ E ++Y + + T T NR +++ YQ + L + S + +KN +
Sbjct 19 VKFLDSLVEDKYYSTKLVKTEKTGNRIDIYFDKGQNYQQVLVKL--SDSIANELGLKNTF 76
Query 167 PSTNLLGSTGIKSQIQADTVNAANSALAYFI-----VAHPMAVVPSNPDRFSRLIPTGST 221
+TNL ++I + A N +YF+ P+A + D+ R+I
Sbjct 77 VTTNLDSLKNTINKIYLEKGFAFNRIKSYFLGLDSSTGKPIASISVIKDK-KRVI----- 130
Query 222 SAVSMSGVTTIPQLAIASRLQEYKDLLGAGGNRYSDWLDTFFASKIEH----VDR-PKLL 276
A + G +P+ I + +E+K G +S L H ++R P+ L
Sbjct 131 DAFKVKGYDKVPKRFIYNLEKEFK-----GKLHHSTQLIALHKRLQNHSFVLLERPPQTL 185
Query 277 FSASQTINVQVIMNQAGPNNFSGL 300
F+ T V + + + N+F G+
Sbjct 186 FTPDST-QVFLFLQKKKNNSFDGV 208
> rag:B739_0600 hypothetical protein
Length=510
Score = 40.0 bits (92), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 47/204 (23%), Positives = 88/204 (43%), Gaps = 30/204 (15%)
Query 113 LEFLDAFFESQFYPSAVASTNNTFNRGNLF------YQIICSDLDSTGSTGDGFPVKNIY 166
++FLD+ E ++Y + + T T NR +++ YQ + L + S + +KN +
Sbjct 19 VKFLDSLVEDKYYSTKLVKTEKTGNRIDIYFDKGQNYQQVLVKL--SDSIANELGLKNTF 76
Query 167 PSTNLLGSTGIKSQIQADTVNAANSALAYFI-----VAHPMAVVPSNPDRFSRLIPTGST 221
+TNL ++I + A N +YF+ P+A + D+ R+I
Sbjct 77 VTTNLDSLKNTINKIYLEKGFAFNRIKSYFLGLDSSTGKPIASISVIKDK-KRVI----- 130
Query 222 SAVSMSGVTTIPQLAIASRLQEYKDLLGAGGNRYSDWLDTFFASKIEH----VDR-PKLL 276
A + G +P+ I + +E+K G +S L H ++R P+ L
Sbjct 131 DAFKVKGYDKVPKRFIYNLEKEFK-----GKLHHSTQLIALHKRLQNHSFVLLERPPQTL 185
Query 277 FSASQTINVQVIMNQAGPNNFSGL 300
F+ T V + + + N+F G+
Sbjct 186 FTPDST-QVFLFLQKKKNNSFDGV 208
> dpp:DICPUDRAFT_37322 hypothetical protein
Length=478
Score = 38.5 bits (88), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 29/110 (26%), Positives = 54/110 (49%), Gaps = 6/110 (5%)
Query 181 IQADTVNAANSALAYFIVAHPMAVVPSNPDRFSRLIPTGSTSAVSMSGVTTIPQLAIASR 240
I+ + A N L++ V V PS +R ++L+ S+VS+ VT +P++AI +
Sbjct 159 IETVKIAAQNPTLSFSCVKFTGLVSPSILERLNQLV-----SSVSVD-VTNLPEVAIKNP 212
Query 241 LQEYKDLLGAGGNRYSDWLDTFFASKIEHVDRPKLLFSASQTINVQVIMN 290
+Q YKD+L + S + E V+R + +F V ++++
Sbjct 213 IQFYKDILASNSEFKSIYTSDEIRQLEEFVNRAERIFIECNKYGVPILLD 262
> fve:101314332 capsid protein VP1-like
Length=421
Score = 37.4 bits (85), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 36/130 (28%), Positives = 58/130 (45%), Gaps = 6/130 (5%)
Query 213 SRLIPTGSTSAVSMSGVTTIPQLAIASRLQEYKDLLGAGGNRYSDWLDTFF--ASKIEHV 270
S + TG + +S + TI QL + ++Q+ + GG RY++ + + F AS +
Sbjct 289 SATVATGLYADLSAATAATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDARL 348
Query 271 DRPKLLFSASQTINVQVIMNQAGPNNFSGLDISGPLGQQGGAIAFNEQLGRRQSYYFSEP 330
RP+ L S IN+ I G G L G +A G S F E
Sbjct 349 QRPEYLGGGSTPINIAPIAQTGGTGAQGTTTPQGNLAAFGTYMA----KGHGFSQSFVEH 404
Query 331 GYLIDMLSIR 340
G++I ++S+R
Sbjct 405 GHVIGLVSVR 414
> tye:THEYE_A1830 heterodisulfide reductase subunit A/methylviologen
reducing hydrogenase subunit delta
Length=744
Score = 37.4 bits (85), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 37/149 (25%), Positives = 60/149 (40%), Gaps = 15/149 (10%)
Query 268 EHVDRPKLLFSASQTINVQVIMNQAGPNNFSGLD-ISGPLGQQG-GAIAFNEQLGRRQSY 325
E VD K+ + V V+ + G+D I + +QG +I R ++Y
Sbjct 17 EAVDVEKIQNIGKNALRVPVVQVHDVFCSREGIDLIKKDIAEQGVNSIVIAGCSPRVKTY 76
Query 326 YFSEPGYLIDMLSIRPVYYWSFVKPDYLNYLGPDYFN---------PIYNDIGYQDVSSA 376
FS PG ++ + +R + W+ P+ DY I + V S
Sbjct 77 EFSFPGCFVERVPLRELAVWTIEAPEEKQLAAEDYIKMGVIKAQKGEIPEPYIIETVKSI 136
Query 377 RIVFNGNAGATSASEPCFNEFRASYDEVL 405
+V G +G T+A+E +A YD VL
Sbjct 137 LVVGGGISGMTAATEAA----KAGYDVVL 161
> hte:Hydth_1101 uroporphyrinogen III synthase HEM4
Length=260
Score = 36.6 bits (83), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 43/100 (43%), Gaps = 11/100 (11%)
Query 403 EVLGQLQGYPMPEVD------GTSYIPLYAYWVQQRTSKLSDGSGSLPESHYYPILFTDM 456
++L Q+ G M E++ G + L+ Y + T +L L E Y+ +LFT
Sbjct 131 KILVQMYGEEMHELEEFLNNKGAKMLKLWLYRYETDTERLDAFIIKLLEGFYHAVLFTSA 190
Query 457 NQVNSPFASKVEDNFFVNMSYAVQKKSLVNKTFATRLSNR 496
QV+ F E N N+S SL NK F + +
Sbjct 191 YQVDYIFKRAKEKNLGKNLS-----SSLNNKVFTVAVGRK 225
> hth:HTH_1109 hemD2; uroporphyrinogen-III synthase
Length=260
Score = 36.6 bits (83), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 43/100 (43%), Gaps = 11/100 (11%)
Query 403 EVLGQLQGYPMPEVD------GTSYIPLYAYWVQQRTSKLSDGSGSLPESHYYPILFTDM 456
++L Q+ G M E++ G + L+ Y + T +L L E Y+ +LFT
Sbjct 131 KILVQMYGEEMHELEEFLNNKGAKMLKLWLYRYETDTERLDAFIIKLLEGFYHAVLFTSA 190
Query 457 NQVNSPFASKVEDNFFVNMSYAVQKKSLVNKTFATRLSNR 496
QV+ F E N N+S SL NK F + +
Sbjct 191 YQVDYIFKRAKEKNLGKNLS-----SSLNNKVFTVAVGRK 225
Lambda K H a alpha
0.318 0.133 0.403 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1081113546656