bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-34_CDS_annotation_glimmer3.pl_2_1
Length=299
Score E
Sequences producing significant alignments: (Bits) Value
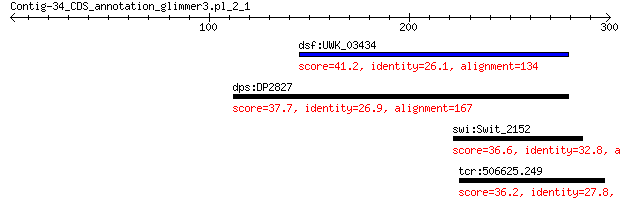
dsf:UWK_03434 ribonucleoside-diphosphate reductase class II (E... 41.2 0.22
dps:DP2827 ribonucleoside-diphosphate reductase 37.7 3.5
swi:Swit_2152 RND efflux system outer membrane lipoprotein 36.6 7.2
tcr:506625.249 RNA pseudouridylate synthase protein 36.2 9.0
> dsf:UWK_03434 ribonucleoside-diphosphate reductase class II
(EC:1.17.4.-)
Length=722
Score = 41.2 bits (95), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 35/139 (25%), Positives = 61/139 (44%), Gaps = 15/139 (11%)
Query 145 DASDFPIPELDSIGMQTQYRCELSAPLMGLNDLLV----PYGYQTSPLDMSVTYGYSPRY 200
D + FPIPE++ + ++T+ ++ +MGL+DLL+ PYG + + Y
Sbjct 323 DCNRFPIPEIEEMTLKTR---KIGMGIMGLHDLLIQLGLPYGSEEGRAQAAEIMSYVREK 379
Query 201 AELKSARDYYEGGFCGTYSTWVTGYDESFLSGWRRNRGSVSVTDYDSIEDLFKCRASLLY 260
AE +S E G Y + Y RRN S+ ++ + C AS
Sbjct 380 AEERSVELAVEKGAFPAYDAKLNKYPA------RRNAALTSIQPTGTVSMIADC-ASGCE 432
Query 261 PIF-VNQWSGTVNDDKLLI 278
P F + ++ D+L++
Sbjct 433 PYFSIVMIKNVMDGDRLIL 451
> dps:DP2827 ribonucleoside-diphosphate reductase
Length=785
Score = 37.7 bits (86), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 45/182 (25%), Positives = 78/182 (43%), Gaps = 28/182 (15%)
Query 112 FTSTTYGMI-IGIYRAIPQLDYSHVGIDRNLFKT---------DASDFPIPELDSIGMQT 161
+ S G I +G + ++DYS +G + KT D + FPIPE+ + ++T
Sbjct 343 YESCNLGSINLGNFVKNDEVDYSRLG---QIVKTATCFLDNVIDCNVFPIPEIGEMTLKT 399
Query 162 QYRCELSAPLMGLNDLL----VPYGYQTSPLDMSVTYGYSPRYAELKSARDYYEGGFCGT 217
+ ++ +MGL+DLL +PYG + L + + E +S + E G
Sbjct 400 R---KIGLGIMGLHDLLIQLAIPYGSEEGRLLAAEVMQFIRDRCEEQSRKLAEEKGAFPA 456
Query 218 YSTWVTGYDESFLSGWRRNRGSVSVTDYDSIEDLFKCRASLLYPIF-VNQWSGTVNDDKL 276
Y + Y RRN S+ ++ + C AS P F + ++ D+L
Sbjct 457 YDAEINLYPA------RRNAALTSIQPTGTVSMIADC-ASGCEPYFSIVMMKHVMDGDRL 509
Query 277 LI 278
L+
Sbjct 510 LM 511
> swi:Swit_2152 RND efflux system outer membrane lipoprotein
Length=503
Score = 36.6 bits (83), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 31/65 (48%), Gaps = 2/65 (3%)
Query 222 VTGYDESF-LSGWRRNRGSVSVTDYDSIEDLFKCRASLLYPIFVNQWSGTVNDDKLLIGS 280
+ DE+F L GWRR G VS DY+ L + + + P N ++ VN +L+G
Sbjct 212 LAAQDETFELVGWRRQAGLVSSLDYEQARQL-RAQTAASIPTIENDYATAVNRIAVLLGE 270
Query 281 VNTCV 285
V
Sbjct 271 APGAV 275
> tcr:506625.249 RNA pseudouridylate synthase protein
Length=842
Score = 36.2 bits (82), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 20/72 (28%), Positives = 39/72 (54%), Gaps = 7/72 (10%)
Query 225 YDESFLSGWRRNRGSVSVTDYDSIEDLFKCRASLLYPIFVNQWSGTVNDDKLLIGSVNTC 284
+ +S +S W R RG D D++++ F C+A+ L V+ W + + + L G ++
Sbjct 706 FSQSAVSKWERVRG-----DLDTMKEFFMCKAASLRNYHVSTWIESCSKVETLAGLLD-- 758
Query 285 VAVRPFSMYGLP 296
+ V+ ++YG P
Sbjct 759 ILVQEINVYGPP 770
Lambda K H a alpha
0.318 0.137 0.409 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 502807620480