bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-39_CDS_annotation_glimmer3.pl_2_1
Length=225
Score E
Sequences producing significant alignments: (Bits) Value
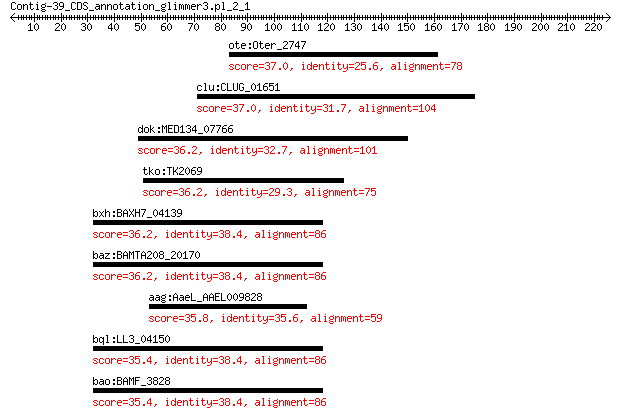
ote:Oter_2747 exopolysaccharide biosynthesis polyprenyl glycos... 37.0 2.6
clu:CLUG_01651 hypothetical protein 37.0 3.0
dok:MED134_07766 hypothetical protein 36.2 4.2
tko:TK2069 cytosolic NiFe-hydrogenase subunit alpha 36.2 4.6
bxh:BAXH7_04139 carbon monoxide dehydrogenase large 36.2 6.0
baz:BAMTA208_20170 xanthine dehydrogenase 36.2 6.0
aag:AaeL_AAEL009828 hypothetical protein 35.8 7.1
bql:LL3_04150 xanthine dehydrogenase 35.4 9.3
bao:BAMF_3828 RBAM_037030; xanthine dehydrogenase (EC:1.17.1.4) 35.4 9.3
> ote:Oter_2747 exopolysaccharide biosynthesis polyprenyl glycosylphosphotransferase
Length=459
Score = 37.0 bits (84), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 20/80 (25%), Positives = 41/80 (51%), Gaps = 4/80 (5%)
Query 83 LEYRG-PDYFNPIYNDIGYQDVPLWRLGYGWKADTVSSLSVAKEPCYNEFRSSYDEVLGS 141
L +RG P Y +++++ ++ +PL+RL W +A+EP + + + D VL S
Sbjct 224 LYFRGVPTYTLELFHEVYWRKIPLYRLNQTWLFQ--EGFQMAREPVFQRLKRTVDIVLAS 281
Query 142 LQATL-TPKASTPLQSYWVQ 160
+ L P + ++W++
Sbjct 282 VGLLLAAPLITLGALAHWLE 301
> clu:CLUG_01651 hypothetical protein
Length=623
Score = 37.0 bits (84), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 33/106 (31%), Positives = 45/106 (42%), Gaps = 17/106 (16%)
Query 71 PVYFWTG--IRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGYGWKADTVSSLSVAKEPCY 128
PV FW+G + PD++E++ F N +DV TVS + A C
Sbjct 385 PVQFWSGSIVFPDFVEFQAKAEFISCTNYKKPEDVT-----------TVSIHNRAIRVCK 433
Query 129 NEFRSSYDEVLGSLQATLTPKASTPLQSYWVQQRDFYLIGLSSNPN 174
F S + G L T + P S RDFYL+ LS+N N
Sbjct 434 EMFEKSKYLIEGRLDRT----RADPYLSQVASSRDFYLVRLSNNDN 475
> dok:MED134_07766 hypothetical protein
Length=304
Score = 36.2 bits (82), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 33/122 (27%), Positives = 57/122 (47%), Gaps = 27/122 (22%)
Query 49 LGREQTYYFKEPGYIF---DMMTIRP-----------VYFWTGIRPDYLE-------YRG 87
+G E +YF GY+F D + ++P V F + +LE YR
Sbjct 187 IGSETNHYFITSGYVFQVSDNVKLKPSVMVKSAFDAPVSFDANVNALFLEKFELGVSYRY 246
Query 88 PDYFNPIYNDIGYQDVPLWRLGYGWKADTVSSLSVAKEPCYNEFRSSYDEVLGSLQATLT 147
D F+ I +G+Q P +R+GY + A T ++A P +E +Y+ +L + +A +
Sbjct 247 DDSFSGI---VGFQATPEFRIGYAYDAVTSEINNIA--PSSHEIIITYN-ILRNKKALRS 300
Query 148 PK 149
P+
Sbjct 301 PR 302
> tko:TK2069 cytosolic NiFe-hydrogenase subunit alpha
Length=428
Score = 36.2 bits (82), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 35/75 (47%), Gaps = 1/75 (1%)
Query 51 REQTYYFKEPGYIFDMMTIRPVYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGY 110
RE+ +E YI DM+ ++ + + PDYL Y GP + Y + L LG
Sbjct 89 REEIQALREVLYIGDMIESHALHLYLLVLPDYLGYSGPLHMIDEYKKEMSIALDLKNLG- 147
Query 111 GWKADTVSSLSVAKE 125
W D + S ++ +E
Sbjct 148 SWMMDELGSRAIHQE 162
> bxh:BAXH7_04139 carbon monoxide dehydrogenase large
Length=776
Score = 36.2 bits (82), Expect = 6.0, Method: Composition-based stats.
Identities = 33/118 (28%), Positives = 50/118 (42%), Gaps = 34/118 (29%)
Query 32 ESAALGQMGGSISFNTVLGREQTYYFKEPGYIFD--MMTIRPVYFWTGIRPDY-LEY-RG 87
E AALGQ+ G +S G +T+ F G + + + T RP+++ G +P+Y +E+
Sbjct 661 EKAALGQVMGGMSMGIAFGGRETFVFDPYGRVLNPQLRTYRPLHY--GEQPEYIIEFVET 718
Query 88 PDYFNP---------------------IYNDIGYQ--DVP-----LWRLGYGWKADTV 117
P P + +G Q +P LWR GWK DTV
Sbjct 719 PHVIAPYGARGAGEHGLLGMPAALGNCLSAALGIQLNQLPLIPEQLWRTQMGWKDDTV 776
> baz:BAMTA208_20170 xanthine dehydrogenase
Length=776
Score = 36.2 bits (82), Expect = 6.0, Method: Composition-based stats.
Identities = 33/118 (28%), Positives = 50/118 (42%), Gaps = 34/118 (29%)
Query 32 ESAALGQMGGSISFNTVLGREQTYYFKEPGYIFD--MMTIRPVYFWTGIRPDY-LEY-RG 87
E AALGQ+ G +S G +T+ F G + + + T RP+++ G +P+Y +E+
Sbjct 661 EKAALGQVMGGMSMGIAFGGRETFVFDPYGRVLNPQLRTYRPLHY--GEQPEYIIEFVET 718
Query 88 PDYFNP---------------------IYNDIGYQ--DVP-----LWRLGYGWKADTV 117
P P + +G Q +P LWR GWK DTV
Sbjct 719 PHVIAPYGARGAGEHGLLGMPAALGNCLSAALGIQLNQLPLIPEQLWRTQMGWKDDTV 776
> aag:AaeL_AAEL009828 hypothetical protein
Length=451
Score = 35.8 bits (81), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 21/59 (36%), Positives = 28/59 (47%), Gaps = 0/59 (0%)
Query 53 QTYYFKEPGYIFDMMTIRPVYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGYG 111
Q Y+ EPGY M+ P T DY EY G D + IY + Q ++RL +G
Sbjct 79 QLYHQPEPGYWMTMILNVPFERKTRETGDYNEYHGDDIHDTIYQAVLKQSYRMFRLFHG 137
> bql:LL3_04150 xanthine dehydrogenase
Length=776
Score = 35.4 bits (80), Expect = 9.3, Method: Composition-based stats.
Identities = 33/118 (28%), Positives = 50/118 (42%), Gaps = 34/118 (29%)
Query 32 ESAALGQMGGSISFNTVLGREQTYYFKEPGYIFD--MMTIRPVYFWTGIRPDY-LEY-RG 87
E AALGQ+ G +S G +T+ F G + + + T RP+++ G +P+Y +E+
Sbjct 661 EKAALGQVMGGMSMGIAFGGRETFDFDPYGRVLNPQLRTYRPLHY--GEQPEYIIEFVET 718
Query 88 PDYFNP---------------------IYNDIGYQ--DVP-----LWRLGYGWKADTV 117
P P + +G Q +P LWR GWK DTV
Sbjct 719 PHVIAPYGARGAGEHGLLGMPAALGNCLSAALGVQLNQLPLIPEQLWRTQMGWKDDTV 776
> bao:BAMF_3828 RBAM_037030; xanthine dehydrogenase (EC:1.17.1.4)
Length=776
Score = 35.4 bits (80), Expect = 9.3, Method: Composition-based stats.
Identities = 33/118 (28%), Positives = 50/118 (42%), Gaps = 34/118 (29%)
Query 32 ESAALGQMGGSISFNTVLGREQTYYFKEPGYIFD--MMTIRPVYFWTGIRPDY-LEY-RG 87
E AALGQ+ G +S G +T+ F G + + + T RP+++ G +P+Y +E+
Sbjct 661 EKAALGQVMGGMSMGIAFGGRETFDFDPYGRVLNPQLRTYRPLHY--GEQPEYIIEFVET 718
Query 88 PDYFNP---------------------IYNDIGYQ--DVP-----LWRLGYGWKADTV 117
P P + +G Q +P LWR GWK DTV
Sbjct 719 PHVIAPYGARGAGEHGLLGMPAALGNCLSAALGVQLNQLPLIPEQLWRTQMGWKDDTV 776
Lambda K H a alpha
0.318 0.135 0.406 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 287955853920