bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-3_CDS_annotation_glimmer3.pl_2_1
Length=349
Score E
Sequences producing significant alignments: (Bits) Value
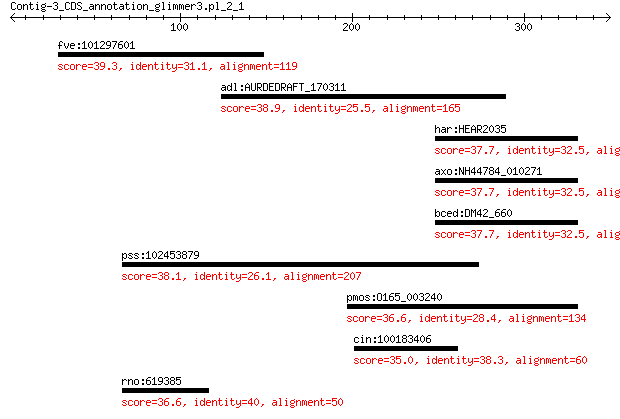
fve:101297601 minor spike protein H-like 39.3 0.27
adl:AURDEDRAFT_170311 hypothetical protein 38.9 1.9
har:HEAR2035 NADH dehydrogenase transmembrane protein (EC:1.6.... 37.7 3.0
axo:NH44784_010271 NADH dehydrogenase (EC:1.6.99.3) 37.7 3.0
bced:DM42_660 pyridine nucleotide-disulfide oxidoreductase fam... 37.7 3.0
pss:102453879 STAB2; stabilin 2 38.1
pmos:O165_003240 NADH dehydrogenase 36.6 7.2
cin:100183406 myosin light polypeptide 6-like 35.0 8.8
rno:619385 Ncoa4; nuclear receptor coactivator 4 36.6
> fve:101297601 minor spike protein H-like
Length=139
Score = 39.3 bits (90), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 56/122 (46%), Gaps = 13/122 (11%)
Query 29 DTNKTNLKIAQMNNDFNAREAQKARDFQLDMWNKENEYNSASSQRERLENAGYNPYMNEA 88
DTN TN IA + + + EAQ+ R+FQ + N + A L +AG NP +
Sbjct 6 DTNATNEAIANQSTETSMAEAQRNREFQERLSNSAYQRQVAD-----LSSAGLNPMLAYI 60
Query 89 QAGTATGMSGTSAASAAGAAPQIPYTPDFQ---SVGVNLASALKMMSEKKKTDIENLNMS 145
+ G A+ SG++ G YT Q S + A A K +EK K + E N++
Sbjct 61 KGGGASTPSGST-----GQVTSAQYTSPIQGAASYRLTSAQAAKTEAEKPKIEAETSNIN 115
Query 146 DL 147
+
Sbjct 116 KI 117
> adl:AURDEDRAFT_170311 hypothetical protein
Length=1380
Score = 38.9 bits (89), Expect = 1.9, Method: Composition-based stats.
Identities = 42/171 (25%), Positives = 75/171 (44%), Gaps = 12/171 (7%)
Query 124 LASALKMMSEKKKTDIENLNMSD--LLRSQIWQNLGATDWRNASPEARAYNLSQGRRAAE 181
LAS +K+ + +L+ D +W+ L A D R A P ++ LS +AA
Sbjct 804 LASQMKIKQPRVFDGRADLDFFDQWCFEVDLWRTLNAIDARWAIPMVSSF-LSD--KAAR 860
Query 182 LGMASLEENLSNQRWSNNLLVANIANSLLDADTKTILNKYLDQQQQAELNVKAANYEYLV 241
M S+ +S++RW L + + D K +L K Q +Q + NV+ + +
Sbjct 861 FFMNSVI--MSDERWDFRKLYDALFDHCFPTDFKLLLRKKFAQARQGQRNVREFARDLKI 918
Query 242 MSGQL----KRQEVNNLIAEEIETYARANGYNLQNRILRETSDGLIRATNN 288
M+ + +RQ + L E + +Y R + + + T D L+ A +
Sbjct 919 MARRFPDVGERQLIQVLF-EGVHSYIRQKWHEFRYNVDEHTYDELVSAAEH 968
> har:HEAR2035 NADH dehydrogenase transmembrane protein (EC:1.6.99.3)
Length=435
Score = 37.7 bits (86), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 27/83 (33%), Positives = 44/83 (53%), Gaps = 2/83 (2%)
Query 248 RQEVNNLIAEEIETYARANGYNLQNRILRETSDGLIRATNNTNFYFGSYYHSRAFNAGAD 307
R EVN L+AE +E + + N +R+T D L+ AT T+ YFG + AF G
Sbjct 73 RPEVNTLMAE-VEGIDQDARQVILNNGMRQTYDTLVLATGATHAYFG-HDEWGAFAPGLK 130
Query 308 AFHDSSILRSRAGSASESYKQSA 330
D++ +R R +A E ++++
Sbjct 131 TLEDATTIRGRILAAFEEAERTS 153
> axo:NH44784_010271 NADH dehydrogenase (EC:1.6.99.3)
Length=435
Score = 37.7 bits (86), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 27/83 (33%), Positives = 44/83 (53%), Gaps = 2/83 (2%)
Query 248 RQEVNNLIAEEIETYARANGYNLQNRILRETSDGLIRATNNTNFYFGSYYHSRAFNAGAD 307
R EVN L+AE +E + + N +R+T D L+ AT T+ YFG + AF G
Sbjct 73 RPEVNTLMAE-VEGIDQDARQVILNNGMRQTYDTLVLATGATHAYFG-HDEWGAFAPGLK 130
Query 308 AFHDSSILRSRAGSASESYKQSA 330
D++ +R R +A E ++++
Sbjct 131 TLEDATTIRGRILAAFEEAERTS 153
> bced:DM42_660 pyridine nucleotide-disulfide oxidoreductase family
protein
Length=435
Score = 37.7 bits (86), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 27/83 (33%), Positives = 44/83 (53%), Gaps = 2/83 (2%)
Query 248 RQEVNNLIAEEIETYARANGYNLQNRILRETSDGLIRATNNTNFYFGSYYHSRAFNAGAD 307
R EVN L+AE +E + + N +R+T D L+ AT T+ YFG + AF G
Sbjct 73 RPEVNTLMAE-VEGIDQDARQVILNNGMRQTYDTLVLATGATHAYFG-HDEWGAFAPGLK 130
Query 308 AFHDSSILRSRAGSASESYKQSA 330
D++ +R R +A E ++++
Sbjct 131 TLEDATTIRGRILAAFEEAERTS 153
> pss:102453879 STAB2; stabilin 2
Length=2557
Score = 38.1 bits (87), Expect = 3.0, Method: Composition-based stats.
Identities = 54/245 (22%), Positives = 96/245 (39%), Gaps = 40/245 (16%)
Query 66 YNSASSQRERL-ENAGYNPYMNEAQAGTATGMSGTSAASAAGAAPQIPYTPD-----FQS 119
Y SA+ + L E AG+N ++NEA +SGTS + + Q D F
Sbjct 994 YGSAAHELSSLSEAAGFNQWVNEASINMM--LSGTSNLTVLVPSLQAIDNMDQEEKAFWM 1051
Query 120 VGVNLASALK---MMSEKKKTDIENLNMSDLLRSQIWQNLGATDWRNASPEARAYNLSQG 176
N+ + LK +M + D++NL+ +D+L + + N N + N+ G
Sbjct 1052 SKNNIQTLLKYHVLMRAYRLADLQNLSSTDMLATSLHNNFLHLSKENENITIEGANIVVG 1111
Query 177 RRAAELG----------------------MASLEENLSNQRWSNNLLVANIANSLLDADT 214
AA G +A LE+ + ++ ++AN + A+T
Sbjct 1112 DIAATNGVIHVIDKVLTPLRGLSGTLPKLLARLEQMPDYSIFRGYVIQYSLANEIEAANT 1171
Query 215 KTI-------LNKYLDQQQQAELNVKAANYEYLVMSGQLKRQEVNNLIAEEIETYARANG 267
T+ + YL ++ L+ Y ++ LK N + E + ++ G
Sbjct 1172 YTVFAPNNDAIESYLRNKKSVTLDEDQIRYHIVLEEKLLKNDMHNGMHRETMLGFSYQVG 1231
Query 268 YNLQN 272
+ LQN
Sbjct 1232 FFLQN 1236
> pmos:O165_003240 NADH dehydrogenase
Length=435
Score = 36.6 bits (83), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 38/136 (28%), Positives = 62/136 (46%), Gaps = 8/136 (6%)
Query 197 SNNLLVANIANSLLDADTKTILNKYLDQQQQAELNVK--AANYEYLVMSGQLKRQEVNNL 254
+N L A + +++D + L Q A L+ A YL + R EVN L
Sbjct 24 ANQLAGAEVDVTIIDRRNHHLFQPLLYQVAGASLSTSEIAWPIRYLFRN----RPEVNTL 79
Query 255 IAEEIETYARANGYNLQNRILRETSDGLIRATNNTNFYFGSYYHSRAFNAGADAFHDSSI 314
+AE +E + + N R+T D L+ AT T+ YFG + AF G D++
Sbjct 80 MAE-VEGIDQDARQVILNNGSRQTYDTLVLATGATHAYFG-HDEWGAFAPGLKTLEDATT 137
Query 315 LRSRAGSASESYKQSA 330
+R R +A E ++++
Sbjct 138 IRGRILAAFEEAERTS 153
> cin:100183406 myosin light polypeptide 6-like
Length=150
Score = 35.0 bits (79), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 23/61 (38%), Positives = 34/61 (56%), Gaps = 1/61 (2%)
Query 201 LVANIANSLLDADTKTILNKYLDQQQQAELNVKAANY-EYLVMSGQLKRQEVNNLIAEEI 259
LV ++ +L D T +NK L ++ ELN K + E+L M Q+KRQ V + I + I
Sbjct 30 LVGSLIRALGDDPTNADVNKVLGNPKKEELNSKTVTFEEFLPMLAQIKRQAVPSNIEDFI 89
Query 260 E 260
E
Sbjct 90 E 90
> rno:619385 Ncoa4; nuclear receptor coactivator 4
Length=622
Score = 36.6 bits (83), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 24/52 (46%), Gaps = 2/52 (4%)
Query 66 YNSASSQRERLENAGYN--PYMNEAQAGTATGMSGTSAASAAGAAPQIPYTP 115
+ S+SS R LE GY P A +GTA +S S PQ PY P
Sbjct 169 HPSSSSVRPFLEKRGYTQVPEQKSASSGTAVSLSEWLLVSKPAIGPQAPYVP 220
Lambda K H a alpha
0.311 0.124 0.344 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 653816197067