bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-11_CDS_annotation_glimmer3.pl_2_1
Length=119
Score E
Sequences producing significant alignments: (Bits) Value
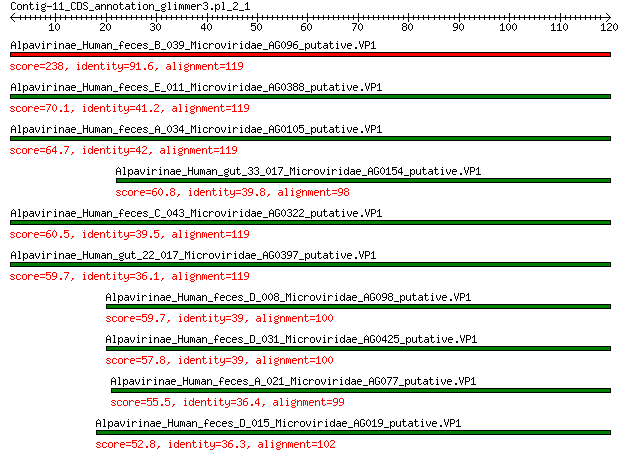
Alpavirinae_Human_feces_B_039_Microviridae_AG096_putative.VP1 238 4e-78
Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1 70.1 1e-16
Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1 64.7 9e-15
Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1 60.8 2e-13
Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1 60.5 3e-13
Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1 59.7 5e-13
Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1 59.7 5e-13
Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1 57.8 3e-12
Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1 55.5 2e-11
Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1 52.8 1e-10
> Alpavirinae_Human_feces_B_039_Microviridae_AG096_putative.VP1
Length=579
Score = 238 bits (608), Expect = 4e-78, Method: Compositional matrix adjust.
Identities = 109/119 (92%), Positives = 116/119 (97%), Gaps = 0/119 (0%)
Query 1 MQSVPSLNLQNNPGRNVSGALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDGWNVL 60
MQ+VP+LNLQNNPGRNVSG+LGYNLRYWQWKSNIDTVHAGFRAGAAYQSW APLDGW VL
Sbjct 461 MQAVPALNLQNNPGRNVSGSLGYNLRYWQWKSNIDTVHAGFRAGAAYQSWAAPLDGWQVL 520
Query 61 TSSGAWSYQSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY 119
T+SG WSYQSMK+RPQQLNSIFVPQ+D+ANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY
Sbjct 521 TASGTWSYQSMKIRPQQLNSIFVPQIDAANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY 579
> Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1
Length=622
Score = 70.1 bits (170), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 49/129 (38%), Positives = 66/129 (51%), Gaps = 15/129 (12%)
Query 1 MQSVPSLNLQNN---PGRNVSGALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDG- 56
M++VP+ L N+ G V+ LGYN RYW WKS ID VH F + WVAP+D
Sbjct 499 MEAVPATTLFNSVLFDGTAVNDFLGYNPRYWPWKSKIDRVHGAFT--TTLKDWVAPIDDD 556
Query 57 ----W--NVLTSSGAWSYQSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQ 110
W + S + S+ KV P L+SIF DS + DQLL N + V+
Sbjct 557 YLHNWFNSKDGKSASISWPFFKVNPNTLDSIFAVVADSIWET---DQLLINCDVSCKVVR 613
Query 111 NLDRNGLPY 119
L ++G+PY
Sbjct 614 PLSQDGMPY 622
> Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1
Length=618
Score = 64.7 bits (156), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 50/131 (38%), Positives = 65/131 (50%), Gaps = 17/131 (13%)
Query 1 MQSVPSLNLQNNP--GRNVSGAL-GYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDG- 56
M+S+P L+L N+ G V+ + GY RY WK++ID V F +SW AP+D
Sbjct 493 MESLPMLSLVNSKAIGDIVARSYAGYVPRYISWKTSIDVVRGAFTD--TLKSWTAPVDSD 550
Query 57 ------WNVLTSSGA--WSYQSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYA 108
V+ G+ SY KV P LN IF VD S DQLLCN F V
Sbjct 551 YMHVFFGEVIPQEGSPILSYTWFKVNPSVLNPIFAVSVDG---SWNTDQLLCNCQFDVKV 607
Query 109 VQNLDRNGLPY 119
+NL +G+PY
Sbjct 608 ARNLSYDGMPY 618
> Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1
Length=614
Score = 60.8 bits (146), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 39/110 (35%), Positives = 56/110 (51%), Gaps = 17/110 (15%)
Query 22 GYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAP-----LDGWNVLTSSGAWS-------YQ 69
GYN RY+ WK+ +D ++ F +SWV+P L GW +S + + Y+
Sbjct 510 GYNPRYFNWKTKLDVINGAFTT--TLKSWVSPVSESLLSGWARFGASDSKTGTKAVLNYK 567
Query 70 SMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY 119
KV P L+ IF + DS + DQLL N Y V+NL R+G+PY
Sbjct 568 FFKVNPSVLDPIFGVKADSTWDT---DQLLVNSYIGCYVVRNLSRDGVPY 614
> Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1
Length=602
Score = 60.5 bits (145), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 47/134 (35%), Positives = 64/134 (48%), Gaps = 20/134 (15%)
Query 1 MQSVPSLNLQNNPGRNVSG------ALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPL 54
M+++P L N + LGY+ RY +K+++D V F SWVAPL
Sbjct 474 MEALPIETLFNEQSTEATALINSIPVLGYSPRYIAYKTSVDWVSGAFET--TLDSWVAPL 531
Query 55 DGWNVLTS------SGA---WSYQSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQ 105
+T SG+ +Y KV P+ L+ IFV + S DQ L NV+F
Sbjct 532 TVNEQITKLLFNPDSGSVYSMNYGFFKVTPRVLDPIFVQECTDTWDS---DQFLVNVSFN 588
Query 106 VYAVQNLDRNGLPY 119
V VQNLD NG+PY
Sbjct 589 VKPVQNLDYNGMPY 602
> Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1
Length=607
Score = 59.7 bits (143), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 43/135 (32%), Positives = 63/135 (47%), Gaps = 21/135 (16%)
Query 1 MQSVPSLNLQNNPGRNVSGAL--GYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAP----- 53
M+++P + N+P ++ GYN RY+ WK+ +D ++ F +SWV+P
Sbjct 478 METLPMTQIFNSPKASIVNLFNAGYNPRYFNWKTKLDVINGAFT--TTLKSWVSPVTESL 535
Query 54 LDGWNVLTSSGA---------WSYQSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNF 104
L GW S +Y+ KV P L+ IF DS S DQLL N
Sbjct 536 LSGWFGFGYSEGDVNSQNKVVLNYKFFKVNPSVLDPIFGVAADSTWDS---DQLLVNSYI 592
Query 105 QVYAVQNLDRNGLPY 119
Y +NL R+G+PY
Sbjct 593 GCYVARNLSRDGVPY 607
> Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1
Length=618
Score = 59.7 bits (143), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 39/111 (35%), Positives = 59/111 (53%), Gaps = 16/111 (14%)
Query 20 ALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDGWNV-------LTSSGAW----SY 68
+GY R++ K+ D V FR+ ++WVAPLD N+ +TSSG +Y
Sbjct 513 TMGYLPRFFDVKTRYDEVLGAFRS--TLKNWVAPLDPANLPQWLQTSVTSSGKLFLNLNY 570
Query 69 QSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY 119
KV P+ L+SIF + DS ++ DQ L + + AV+N D +G+PY
Sbjct 571 GFFKVNPRVLDSIFNVKCDS---TIDTDQFLTTLYMDIKAVRNFDYDGMPY 618
> Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1
Length=614
Score = 57.8 bits (138), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 39/111 (35%), Positives = 58/111 (52%), Gaps = 16/111 (14%)
Query 20 ALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDGWNV-------LTSSGAWS----Y 68
+GY R++ K+ D V FR+ ++WVAPLD V +TSSG + Y
Sbjct 509 TMGYLPRFFDVKTRYDEVLGAFRS--TLKNWVAPLDPSYVSKWLQSSVTSSGKLALNLNY 566
Query 69 QSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY 119
KV P+ L+SIF + DS ++ DQ L + + AV+N D +G+PY
Sbjct 567 GFFKVNPRVLDSIFNVKCDS---TIDTDQFLTALYMDIKAVRNFDYDGMPY 614
> Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1
Length=622
Score = 55.5 bits (132), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/110 (33%), Positives = 57/110 (52%), Gaps = 16/110 (15%)
Query 21 LGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDG-----W--NVLTSSGA----WSYQ 69
+GY RY WK++ID + FR+ ++WVAP+D W NV ++ A ++Y
Sbjct 518 IGYVPRYVNWKTDIDEIFGAFRSSE--KTWVAPIDADFITNWVKNVADNASAVQSLFNYN 575
Query 70 SMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY 119
KV P L+ IF + DS ++ D + + V+NLD +G+PY
Sbjct 576 WFKVNPAILDDIFAVKADS---TMDTDTFKTRMMMSIKCVRNLDYSGMPY 622
> Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1
Length=584
Score = 52.8 bits (125), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 37/109 (34%), Positives = 53/109 (49%), Gaps = 12/109 (11%)
Query 18 SGALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPL--DGW-NVL----TSSGAWSYQS 70
+ +GY RY+ WK++ID V F + WVAP+ + W N+L T +Y
Sbjct 481 TTTVGYLPRYYAWKTSIDYVLGAFTTTE--KEWVAPITPELWSNMLKPLGTKGTGINYNF 538
Query 71 MKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY 119
KV P L+ IF DS + D L N F + +NLD +G+PY
Sbjct 539 FKVNPSILDPIFAVNADSYWDT---DTFLINAAFDIRVARNLDYDGMPY 584
Lambda K H a alpha
0.318 0.131 0.423 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 6810960