bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-15_CDS_annotation_glimmer3.pl_2_4
Length=102
Score E
Sequences producing significant alignments: (Bits) Value
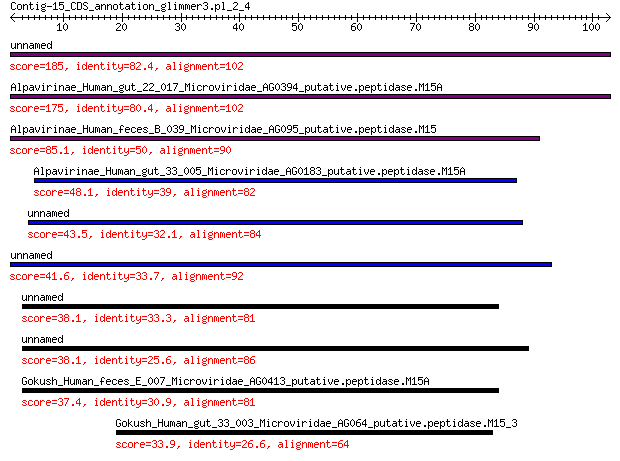
unnamed protein product 185 4e-63
Alpavirinae_Human_gut_22_017_Microviridae_AG0394_putative.pepti... 175 4e-59
Alpavirinae_Human_feces_B_039_Microviridae_AG095_putative.pepti... 85.1 6e-24
Alpavirinae_Human_gut_33_005_Microviridae_AG0183_putative.pepti... 48.1 3e-10
unnamed protein product 43.5 2e-08
unnamed protein product 41.6 8e-08
unnamed protein product 38.1 1e-06
unnamed protein product 38.1 1e-06
Gokush_Human_feces_E_007_Microviridae_AG0413_putative.peptidase... 37.4 3e-06
Gokush_Human_gut_33_003_Microviridae_AG064_putative.peptidase.M... 33.9 7e-05
> unnamed protein product
Length=102
Score = 185 bits (470), Expect = 4e-63, Method: Compositional matrix adjust.
Identities = 84/102 (82%), Positives = 95/102 (93%), Gaps = 0/102 (0%)
Query 1 MNPELMKFVEWLLHRNIHFTVTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFSVDEF 60
MNPELMKFVEW+LHRN+HFTVTSAFRTE+QN+ CNG+KTSQHLTGDAIDLKP+ SVD F
Sbjct 1 MNPELMKFVEWILHRNVHFTVTSAFRTEEQNDACNGSKTSQHLTGDAIDLKPVDSSVDGF 60
Query 61 ISMIKDSPFKFDQLIKYRSFIHISFARGRKPRLMELNFTDRK 102
+ MIK SPFKFDQ+IKYR+F+H+SFARGRKPR MELNFTDRK
Sbjct 61 LLMIKGSPFKFDQIIKYRTFVHVSFARGRKPRQMELNFTDRK 102
> Alpavirinae_Human_gut_22_017_Microviridae_AG0394_putative.peptidase.M15A
Length=102
Score = 175 bits (443), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 82/102 (80%), Positives = 90/102 (88%), Gaps = 0/102 (0%)
Query 1 MNPELMKFVEWLLHRNIHFTVTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFSVDEF 60
MNPELMKFVEWLL RN+HFTVTSAFRT++QN CNG+ SQHLTGDAIDLKPL FSVD
Sbjct 1 MNPELMKFVEWLLRRNLHFTVTSAFRTKEQNEACNGSDHSQHLTGDAIDLKPLDFSVDLL 60
Query 61 ISMIKDSPFKFDQLIKYRSFIHISFARGRKPRLMELNFTDRK 102
+SMIK S FKFDQLIKYR+F+HISFARGR PR MELNFT+RK
Sbjct 61 VSMIKGSSFKFDQLIKYRTFVHISFARGRNPRQMELNFTNRK 102
> Alpavirinae_Human_feces_B_039_Microviridae_AG095_putative.peptidase.M15
Length=102
Score = 85.1 bits (209), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 45/95 (47%), Positives = 60/95 (63%), Gaps = 5/95 (5%)
Query 1 MNPELMKFVEWLLHRNIHFTVTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPL----YFS 56
MN LM F E+LL+ N+HF+VTSA RT +QN G SQHL G+A+D+KP +
Sbjct 1 MNYTLMHFFEYLLYSNVHFSVTSARRTPEQNKAAGGVPNSQHLVGEAVDIKPYGSTTFNK 60
Query 57 VDEFISMIKDSPFKFDQLIKYRSFIHISF-ARGRK 90
+ E I D+ FDQLI Y +FIH+SF +R R+
Sbjct 61 LLEMIHFFSDNVSPFDQLIIYPTFIHVSFCSRNRR 95
> Alpavirinae_Human_gut_33_005_Microviridae_AG0183_putative.peptidase.M15A
Length=131
Score = 48.1 bits (113), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 32/92 (35%), Positives = 47/92 (51%), Gaps = 15/92 (16%)
Query 5 LMKFVEWLLH--RNIH---FTVTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFSVDE 59
L++ W+L R ++ VTS FRT N G TSQH+ G A DL+ D+
Sbjct 33 LLELCRWILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAADLR-----CDD 87
Query 60 ---FISMIKDSPFKFDQLIKY--RSFIHISFA 86
+I +S FDQLI Y + F+H+S++
Sbjct 88 PKALFDLIAESDLPFDQLIYYQKKKFVHVSYS 119
> unnamed protein product
Length=147
Score = 43.5 bits (101), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 27/88 (31%), Positives = 42/88 (48%), Gaps = 4/88 (5%)
Query 4 ELMKFVEWLLHR-NIHFTVTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFSVDEFIS 62
+L F+E L + VTS FR NN G S HL G A D++ ++ + +
Sbjct 40 QLCLFLEQLRSQLGCAVIVTSGFRNVSVNNSVGGVLNSDHLYGLAADIRVKGYTPTKLCN 99
Query 63 MIKDSPF---KFDQLIKYRSFIHISFAR 87
I+ P + Q+I Y +F+H+S R
Sbjct 100 FIRSIPLLNVQVGQVIIYPTFLHVSINR 127
> unnamed protein product
Length=130
Score = 41.6 bits (96), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 31/94 (33%), Positives = 46/94 (49%), Gaps = 4/94 (4%)
Query 1 MNPELMKFVEWL-LHRNIHFTVTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFSVDE 59
++ EL+ +E + H N VTS +RT + N + G K SQH G A D+K E
Sbjct 36 VDTELIDVLEDIRAHFNKPVIVTSGYRTPEYNTKIGGVKNSQHTKGTAADIKVSGIPAKE 95
Query 60 FISMIKDS-PFKFDQLIKYRSFIHISFARGRKPR 92
+K+ P K+ + Y +F HI R +K R
Sbjct 96 VQKYLKNKYPNKYG-IGSYLNFTHIDI-RAKKAR 127
> unnamed protein product
Length=128
Score = 38.1 bits (87), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/85 (32%), Positives = 41/85 (48%), Gaps = 8/85 (9%)
Query 3 PELMKFVEWLL-HRNIHFTVTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFS---VD 58
PEL+ +E + H N + S +RT N + GA+ S H G A D++ + V
Sbjct 36 PELLGILETIREHFNEPVIINSGYRTPAWNKKVGGAENSYHCKGMAADIRVKGHTSKEVA 95
Query 59 EFISMIKDSPFKFDQLIKYRSFIHI 83
E+ S I K +I+Y +F HI
Sbjct 96 EYASKI----MKAGGVIRYTNFTHI 116
> unnamed protein product
Length=130
Score = 38.1 bits (87), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 22/87 (25%), Positives = 38/87 (44%), Gaps = 1/87 (1%)
Query 3 PELMKFVEWLLHR-NIHFTVTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFSVDEFI 61
P L+ +E + N + S +RT + N + GAK S H+ G A D+ + E
Sbjct 37 PNLIDVLEIIRESVNAPVIINSGYRTPEWNAKVGGAKCSYHVKGMAADIAVKGHTTKEVA 96
Query 62 SMIKDSPFKFDQLIKYRSFIHISFARG 88
+ +I+Y +F+H+ G
Sbjct 97 EIASRILGNHGGVIRYTNFVHVDVREG 123
> Gokush_Human_feces_E_007_Microviridae_AG0413_putative.peptidase.M15A
Length=127
Score = 37.4 bits (85), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 39/82 (48%), Gaps = 2/82 (2%)
Query 3 PELMKFVEWLL-HRNIHFTVTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFSVDEFI 61
PEL++ +E + H + + S +RT N + GA S H G A D+ S E +
Sbjct 36 PELLEILEIIRNHFDAPVIINSGYRTPSWNAKVGGAPNSYHCKGMAADIVVKGHSSKE-V 94
Query 62 SMIKDSPFKFDQLIKYRSFIHI 83
+ DS +I+Y +F HI
Sbjct 95 AKYADSIMVTGGVIRYTNFTHI 116
> Gokush_Human_gut_33_003_Microviridae_AG064_putative.peptidase.M15_3
Length=130
Score = 33.9 bits (76), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 17/64 (27%), Positives = 29/64 (45%), Gaps = 0/64 (0%)
Query 19 FTVTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFSVDEFISMIKDSPFKFDQLIKYR 78
+TS +RT N + GAK S H G A D++ + E ++ +I Y
Sbjct 55 IVITSGYRTVSHNKKVGGAKYSYHTRGMAADIRANGITPKELAKVLSSIVPNSGGIIVYD 114
Query 79 SFIH 82
+++H
Sbjct 115 NWVH 118
Lambda K H a alpha
0.325 0.138 0.415 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 5159970