bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
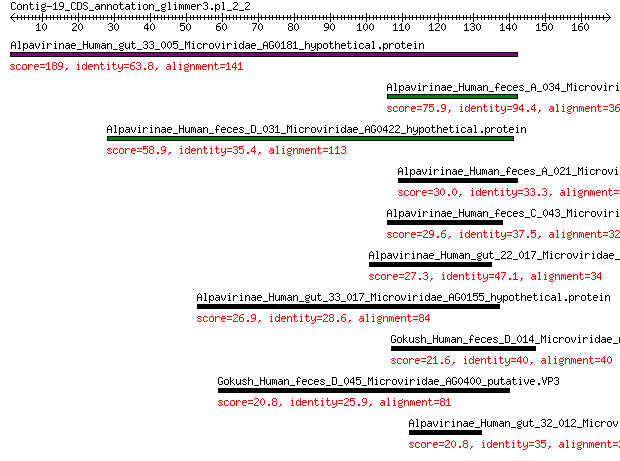
Query= Contig-19_CDS_annotation_glimmer3.pl_2_2
Length=167
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_gut_33_005_Microviridae_AG0181_hypothetical.p... 189 3e-62
Alpavirinae_Human_feces_A_034_Microviridae_AG0103_hypothetical.... 75.9 4e-20
Alpavirinae_Human_feces_D_031_Microviridae_AG0422_hypothetical.... 58.9 2e-13
Alpavirinae_Human_feces_A_021_Microviridae_AG078_hypothetical.p... 30.0 0.002
Alpavirinae_Human_feces_C_043_Microviridae_AG0323_hypothetical.... 29.6 0.002
Alpavirinae_Human_gut_22_017_Microviridae_AG0398_hypothetical.p... 27.3 0.015
Alpavirinae_Human_gut_33_017_Microviridae_AG0155_hypothetical.p... 26.9 0.059
Gokush_Human_feces_D_014_Microviridae_AG029_putative.VP1 21.6 4.9
Gokush_Human_feces_D_045_Microviridae_AG0400_putative.VP3 20.8 6.3
Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1 20.8 9.5
> Alpavirinae_Human_gut_33_005_Microviridae_AG0181_hypothetical.protein
Length=205
Score = 189 bits (480), Expect = 3e-62, Method: Compositional matrix adjust.
Identities = 90/141 (64%), Positives = 108/141 (77%), Gaps = 0/141 (0%)
Query 1 MKKDVIIRGFARYSLNRDNQLHNCTCRGVHSAAVALSGVLREQSPVEDFLFEDNFDGSRT 60
M+KD IIRGFA+YSLNRD H+ +CR V ++ + G+L E +PV DFLF+ N DGS T
Sbjct 41 MEKDAIIRGFAKYSLNRDISDHSQSCRCVMTSEMLAQGILPEPNPVGDFLFDHNADGSVT 100
Query 61 FVSDYSVLSGQAAIDQMNPTQLRRYLNGLRPSSSPYTHRYNDDFLLEYCKDRNIQSYTEM 120
F SDY +L GQ AID MN QLRRY+N L P SS YT YNDDFLL+YCKDRNIQS TEM
Sbjct 101 FCSDYGILFGQKAIDNMNQVQLRRYMNSLVPRSSNYTRNYNDDFLLDYCKDRNIQSATEM 160
Query 121 QAWLEHLISEGQSLENDVAAY 141
+WL+HL+SEGQSLE+D+ AY
Sbjct 161 ASWLDHLLSEGQSLESDLQAY 181
> Alpavirinae_Human_feces_A_034_Microviridae_AG0103_hypothetical.protein
Length=62
Score = 75.9 bits (185), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 34/36 (94%), Positives = 35/36 (97%), Gaps = 0/36 (0%)
Query 106 LEYCKDRNIQSYTEMQAWLEHLISEGQSLENDVAAY 141
+EYCKDRNIQSYTEMQAWLEHLISEGQSLE DVAAY
Sbjct 1 MEYCKDRNIQSYTEMQAWLEHLISEGQSLEGDVAAY 36
> Alpavirinae_Human_feces_D_031_Microviridae_AG0422_hypothetical.protein
Length=150
Score = 58.9 bits (141), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 40/122 (33%), Positives = 63/122 (52%), Gaps = 17/122 (14%)
Query 28 GVHSAAVALSGVLREQSPVEDFLFEDNFDGSRTFVSDYSVLSGQAAIDQMNPTQLRR--- 84
+ +A V S + + PVE + + DG +VSD ++L MN +LR
Sbjct 15 ALDTARVLKSAIYCQVGPVEMLRYVKDDDGVIRYVSDVNLL--------MNAERLRNQIG 66
Query 85 ---YLN---GLRPSSSPYTHRYNDDFLLEYCKDRNIQSYTEMQAWLEHLISEGQSLENDV 138
YLN G++P SPY ++Y D+ L K R IQ+ +E+ AW+E L S G S+ +++
Sbjct 67 EESYLNLIRGIQPKKSPYDNKYTDEQLFTAIKSRFIQTPSEVLAWIESLGSAGDSIRSEL 126
Query 139 AA 140
A
Sbjct 127 DA 128
> Alpavirinae_Human_feces_A_021_Microviridae_AG078_hypothetical.protein
Length=63
Score = 30.0 bits (66), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 20/33 (61%), Gaps = 0/33 (0%)
Query 109 CKDRNIQSYTEMQAWLEHLISEGQSLENDVAAY 141
K RN+QS+ E++AW E L + + ++ + Y
Sbjct 4 IKPRNVQSHAELKAWSEFLTDKAKEIQTEYETY 36
> Alpavirinae_Human_feces_C_043_Microviridae_AG0323_hypothetical.protein
Length=59
Score = 29.6 bits (65), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 12/32 (38%), Positives = 21/32 (66%), Gaps = 0/32 (0%)
Query 106 LEYCKDRNIQSYTEMQAWLEHLISEGQSLEND 137
+E K R +QS +E++AWLE L+ + + +D
Sbjct 1 METIKSRYLQSPSEVRAWLETLVDKADVVRSD 32
> Alpavirinae_Human_gut_22_017_Microviridae_AG0398_hypothetical.protein
Length=69
Score = 27.3 bits (59), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 7/41 (17%)
Query 101 NDDFLLEYCKDRNIQSYTEMQAW-------LEHLISEGQSL 134
+DD LL + R+IQ+ +E+ AW EHL S+ Q L
Sbjct 5 SDDDLLATVRSRHIQAPSEIIAWSKELSAYAEHLESQAQEL 45
> Alpavirinae_Human_gut_33_017_Microviridae_AG0155_hypothetical.protein
Length=171
Score = 26.9 bits (58), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 24/100 (24%), Positives = 49/100 (49%), Gaps = 27/100 (27%)
Query 53 DNFDGSRT--FVSDYSV------LSGQAAID--------QMNPTQLRRYLNGLRPSSSPY 96
D+ D SR + SD + L+ +A +D + +P+Q+++ ++ +
Sbjct 54 DDTDASRPVRYTSDIRLILHNKDLASRAGVDVASKFGQSKQSPSQIQQIMDTM------- 106
Query 97 THRYNDDFLLEYCKDRNIQSYTEMQAWLEHLISEGQSLEN 136
+D+ LL + R IQS +E+ AW + L + ++LE+
Sbjct 107 ----SDEDLLATVRSRYIQSPSEILAWSKELSAYAENLES 142
> Gokush_Human_feces_D_014_Microviridae_AG029_putative.VP1
Length=578
Score = 21.6 bits (44), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 16/46 (35%), Positives = 23/46 (50%), Gaps = 6/46 (13%)
Query 107 EYCKDRNIQSYT-----EMQAWLEHLISEGQSLE-NDVAAYQAAKL 146
E+ +D+N+Q + Q HLI EG S+ + AA Q A L
Sbjct 169 EWFRDQNLQDPVLIDKGDSQTNGRHLIPEGNSITFENQAALQGANL 214
> Gokush_Human_feces_D_045_Microviridae_AG0400_putative.VP3
Length=109
Score = 20.8 bits (42), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 21/87 (24%), Positives = 33/87 (38%), Gaps = 18/87 (21%)
Query 59 RTFVSDYSVLSGQAAI------DQMNPTQLRRYLNGLRPSSSPYTHRYNDDFLLEYCKDR 112
R V D SVLSG+ AI NP + LN S S + D
Sbjct 18 RLSVGDTSVLSGRQAIYGDFSALPQNPVDMINVLNSAEQSFS------------QLPADE 65
Query 113 NIQSYTEMQAWLEHLISEGQSLENDVA 139
+ + WL +L++ + + D++
Sbjct 66 KAAFNNDYRVWLANLLNSASNPDVDIS 92
> Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1
Length=604
Score = 20.8 bits (42), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 7/20 (35%), Positives = 13/20 (65%), Gaps = 0/20 (0%)
Query 112 RNIQSYTEMQAWLEHLISEG 131
RNI + + +Q WL+ ++ G
Sbjct 298 RNIAAQSHIQRWLDLALAGG 317
Lambda K H a alpha
0.318 0.132 0.388 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 11566464