bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-19_CDS_annotation_glimmer3.pl_2_3
Length=365
Score E
Sequences producing significant alignments: (Bits) Value
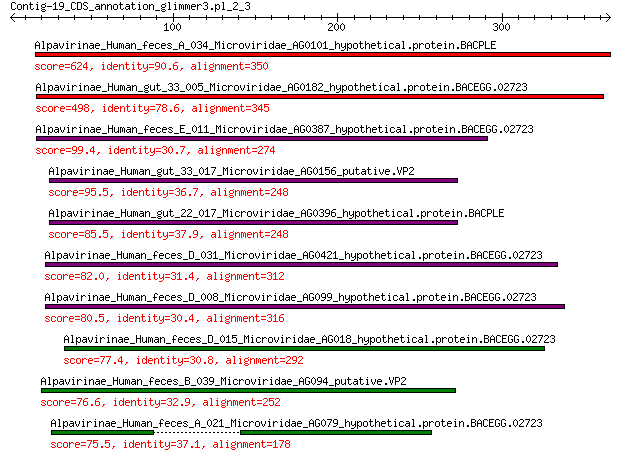
Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.... 624 0.0
Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.p... 498 2e-178
Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.... 99.4 4e-25
Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2 95.5 5e-24
Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.p... 85.5 3e-20
Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.... 82.0 5e-19
Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.p... 80.5 2e-18
Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.p... 77.4 2e-17
Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2 76.6 2e-17
Alpavirinae_Human_feces_A_021_Microviridae_AG079_hypothetical.p... 75.5 7e-17
> Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.protein.BACPLE
Length=365
Score = 624 bits (1610), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 317/350 (91%), Positives = 339/350 (97%), Gaps = 0/350 (0%)
Query 16 VVNAIGNNRQGSKNRKHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARL 75
VVNAIGNNRQGSKNRKHQL+MQ+IQNEWASSESQKSRDFAKSMFDA+NEWNSAK+QRARL
Sbjct 16 VVNAIGNNRQGSKNRKHQLQMQKIQNEWASSESQKSRDFAKSMFDASNEWNSAKSQRARL 75
Query 76 EEAGLNPYLMMNggsagtasstsantvsgasgsggtPYQYTPTNIIGDVASFAGAMKSLS 135
EEAGLNPYLMMNGGSAGTA STSA SG+SGSGG PYQYTPTN+IGDVAS+AGAMKSLS
Sbjct 76 EEAGLNPYLMMNGGSAGTAQSTSATASSGSSGSGGMPYQYTPTNVIGDVASYAGAMKSLS 135
Query 136 EARKTNTESDLLGKYGDSDYSSRIANTEADTYFKQRQSDVATAQRANLLLSSKAQEIMNM 195
+ARK+ TE+DLLG+YGDSDYSSRIANTEADTYFKQRQSDVATAQ+ANLLLS++AQ++MNM
Sbjct 136 DARKSGTEADLLGRYGDSDYSSRIANTEADTYFKQRQSDVATAQKANLLLSAEAQQVMNM 195
Query 196 YLPQEKQIELSTLGAQYWNLIRDGSIKEEQAKNLLATRLEIAARTAGQHISNKVARSTAD 255
YLPQEKQI+LSTLGAQYWN+IRDGSIKEEQAKNLLATRLEI ARTAGQHISNKVARSTAD
Sbjct 196 YLPQEKQIQLSTLGAQYWNMIRDGSIKEEQAKNLLATRLEIEARTAGQHISNKVARSTAD 255
Query 256 SIIDATNTAKMNEAAYNRGYSQFSNDVGFRTGKMDRWLQDPVKARWDRGINNAGEFIEGL 315
SIIDATNTAKMNEAAYNRGYSQFSNDVG+RTGKMDRW QDPVKARWDRGINNAG+FI+GL
Sbjct 256 SIIDATNTAKMNEAAYNRGYSQFSNDVGYRTGKMDRWSQDPVKARWDRGINNAGKFIDGL 315
Query 316 SNIVGSVTKFGFLRNANQRLKEDKRQFDRGTVDIFDDNKGYSWTRQHRGR 365
SNIVGSVTKFGFL+NAN LKE KRQFDRGTVDIFDDNKGYSWTRQHRGR
Sbjct 316 SNIVGSVTKFGFLKNANNSLKESKRQFDRGTVDIFDDNKGYSWTRQHRGR 365
> Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.protein.BACEGG.02723
Length=354
Score = 498 bits (1283), Expect = 2e-178, Method: Compositional matrix adjust.
Identities = 271/346 (78%), Positives = 300/346 (87%), Gaps = 13/346 (4%)
Query 17 VNAIGNNRQGSKNRKHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLE 76
VN IGNN+QG KNRKHQLEMQ+IQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLE
Sbjct 17 VNQIGNNKQGDKNRKHQLEMQQIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLE 76
Query 77 EAGLNPYLMMNggsagtasstsantvsgasgsggtPYQYTPTNIIGDVASFAGAMKSLSE 136
AGLNPYLMMNGGSAGTASSTSA+TVSGASGSGGTPYQYTPTN+IGDVAS+A AMKS+S+
Sbjct 77 AAGLNPYLMMNGGSAGTASSTSASTVSGASGSGGTPYQYTPTNMIGDVASYASAMKSMSD 136
Query 137 ARKTNTESDLLGKYGDSDYSSRIANTEADTYFKQRQSDVATAQRANLLLSSKAQEIMNMY 196
ARKTNTESDLL KYG Y S+I T ADTYF QRQ+DVA AQ+ANLLL + AQE++NMY
Sbjct 137 ARKTNTESDLLDKYGVPTYESQIGKTMADTYFTQRQADVAIAQKANLLLRNDAQEVLNMY 196
Query 197 LPQEKQIELSTLGAQYWNLIRDGSIKEEQAKNLLATRLEIAARTAGQHISNKVARSTADS 256
LP+EK+I+L GAQYWN+IR+G I EEQAKNL+A+RLEI ART GQHISNK+A+STADS
Sbjct 197 LPEEKRIQLQMNGAQYWNMIREGVISEEQAKNLIASRLEIEARTQGQHISNKIAKSTADS 256
Query 257 IIDATNTAKMNEAAYNRGYSQFSNDVGFRTGKMDRWLQDPVKARWDRGINNAGEFIEGLS 316
IIDAT TAK NEAA+NRGYSQFSNDVGFRTGKMDRWLQDPVKARWDRGINNAG+FI+GLS
Sbjct 257 IIDATRTAKENEAAFNRGYSQFSNDVGFRTGKMDRWLQDPVKARWDRGINNAGKFIDGLS 316
Query 317 NIVGSVTKFG-FLRNANQRLKEDKRQFDRGTVDIFDDNKGYSWTRQ 361
N VGS+ KFG FLR R ++ FDDN+GYS TRQ
Sbjct 317 NAVGSLLKFGSFLREG------------RSVIEQFDDNQGYSSTRQ 350
> Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.protein.BACEGG.02723
Length=410
Score = 99.4 bits (246), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 84/305 (28%), Positives = 149/305 (49%), Gaps = 35/305 (11%)
Query 17 VNAIGNNRQGSKNRKHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLE 76
V+ +GN + + +++ R+ NE+ + E++K+R + M++ TN+WNS KN R RL+
Sbjct 60 VSGVGNIFGSALSNSQNMKINRMNNEFNAREAEKARQYQSEMWNKTNDWNSPKNVRKRLQ 119
Query 77 EAGLNPYLMMNggsagtasstsantvsgasgsggtPYQYTPTNIIGDVASFAGAMKSLSE 136
EAG NPYL ++ + GTA S +++ + A+ Q P G + + A++ +
Sbjct 120 EAGYNPYLGLDSSNVGTAQSAGSSSPASAAPP----IQNNPIQFDGFQNALSTAIQMSNS 175
Query 137 ARKTNTES-DLLGKYGDSDYSSRIANTEADTY---------------------FKQRQSD 174
+ +N E+ +L G+ G +D + + D Y F Q +
Sbjct 176 TKVSNAEANNLQGQKGLADAQAAATLSGIDWYKFTPEYRAWLQTTGMARAQLSFNTDQQN 235
Query 175 ---------VATAQRANLLLSSKAQEIMNMYLPQEKQIELSTLGAQYWNLIRDGSIKEEQ 225
+ AQR ++LLS++A+ I+N YL + ++L + Q + G + +Q
Sbjct 236 LENMKWVNKIQRAQRTDILLSNQAKGIINKYLDTSQSLQLKLMANQSFQAFASGRLSLQQ 295
Query 226 AKNLLATRLEIAARTAGQHISNKVARSTADSIIDATNTAKMNEAAYNRGYSQFSNDVGFR 285
K + +L A T G+ ISNK+A TAD +I A + Y+RGY+ ++ + G
Sbjct 296 CKTEVTKQLMNMAETEGKKISNKIASETADQLIGALQWQYSADEMYSRGYAGYAREAGKS 355
Query 286 TGKMD 290
GK D
Sbjct 356 RGKGD 360
> Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2
Length=353
Score = 95.5 bits (236), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 91/284 (32%), Positives = 148/284 (52%), Gaps = 40/284 (14%)
Query 25 QGSKNRKHQLE-------MQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEE 77
Q S+NR++ E + ++ N++ + + RDF ++M++ N +N+A QR RLEE
Sbjct 20 QNSQNRQNIKETNQMNYKINQMNNQFNERMAMQQRDFQENMWNKENTYNTASAQRQRLEE 79
Query 78 AGLNPYLMMNggsagtasstsantvsgasgsgg-tPYQYTPTNI---IGDV--------- 124
AGLNPYLMMNGGS+G + S + +SG+ P+Q + I IG V
Sbjct 80 AGLNPYLMMNGGSSGVSQSAGTGASASSSGTAVFQPFQADFSGIQQAIGSVFQSQVRQAQ 139
Query 125 ---------ASFAGAMKSLSE---ARKTNTESDLLGKYGDS----DYSSRIANTEADTYF 168
+ A AM++LS+ ++ T + L G + YS + + + F
Sbjct 140 VSQMQGQRNLADAQAMQALSQVDWSKMTKETREYLKATGLARARLGYSKEMQELD-NMAF 198
Query 169 KQRQSDVATAQRANLLLSSKAQEIMNMYLPQEKQIELSTLGAQYWNLIRDGSIKEEQAKN 228
R + AQ + LL + A+ ++N YL Q++Q +L+ + Y+N + G + QAK
Sbjct 199 AGR---LLQAQGTSQLLEADAKTVLNRYLDQQQQADLNVKASVYYNQMSQGHLNYNQAKK 255
Query 229 LLATRLEIAARTAGQHISNKVARSTADSIIDATNTAKMNEAAYN 272
++A + AR GQ +SNKVA +TADS+I ATN A + A ++
Sbjct 256 VIADEILTYARIKGQKLSNKVAEATADSLIRATNAANRSNAEFD 299
> Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.protein.BACPLE
Length=386
Score = 85.5 bits (210), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 94/281 (33%), Positives = 152/281 (54%), Gaps = 34/281 (12%)
Query 25 QGSKNRKHQLE-------MQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEE 77
Q S+NR++ E + ++ N++ + + R++ ++M++ N +N+A QR RLEE
Sbjct 20 QNSQNRQNVRETNQMNYKINQMNNQFNERMAIQQRNWQENMWNKENAYNTASAQRQRLEE 79
Query 78 AGLNPYLMMNggsagtasstsantvsgas-gsggtPYQYTPTNI---IGDV--------- 124
AGLNPYLMMNGGSAG A S + + +S + P+Q + I IG++
Sbjct 80 AGLNPYLMMNGGSAGVAQSAGTGSAASSSGNAVMQPFQADYSGIGSSIGNIFQYELMQSE 139
Query 125 -ASFAGAMKSLSEARKTNTESDL-LGKYGDSD----YSSRIANTEADTYFKQRQSD---- 174
+ GA + L++A+ T S++ GK D S+ +A + +Q+++D
Sbjct 140 KSQLQGA-RQLADAKAMETLSNIDWGKLTDETRGFLKSTGLARAQLGYAKEQQEADNMAM 198
Query 175 ---VATAQRANLLLSSKAQEIMNMYLPQEKQIELSTLGAQYWNLIRDGSIKEEQAKNLLA 231
V AQR+ +LL ++A+ I+N YL Q +Q++LS A Y+ + G + +AK LA
Sbjct 199 TGLVLRAQRSGMLLDNEAKGILNKYLDQHQQLDLSVKAADYYQRMAAGYVSYAEAKKALA 258
Query 232 TRLEIAARTAGQHISNKVARSTADSIIDATNTAKMNEAAYN 272
AART GQ+ISN+VA A+S I A A + AAY+
Sbjct 259 EEALAAARTRGQNISNEVASRIAESQIAANIAANESSAAYH 299
> Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.protein.BACEGG.02723
Length=418
Score = 82.0 bits (201), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 98/378 (26%), Positives = 161/378 (43%), Gaps = 66/378 (17%)
Query 22 NNRQGSKNRKHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEW-----------NSAKN 70
+N+ ++ K E+ +I NE+ +SE+ K+RDF S +A+ +W N
Sbjct 27 SNKSVKRSIKAAKEINQINNEFNASEALKNRDFQTSEREASQQWNLDQWNRENAYNDPSA 86
Query 71 QRARLEEAGLNPYLM-MNggsagtasstsantvsgasgsggtPYQYTPTNIIGDVASFAG 129
QRAR+E AG NPY M ++ GS T+ + S+ ++ + TP T D + A
Sbjct 87 QRARMEAAGFNPYNMNIDPGSGSTSGAQSSPGSGSSATASHTPSLPAYTGYAADFQNVAS 146
Query 130 AMKSLSEARKTNTESDLLGKYGD----SDYSSRI-ANTE--ADTYFKQRQSD-------- 174
+ + A + ++ L YGD +D S+I N+E D Y RQ++
Sbjct 147 GIAQIGNAVASGIDARLTSAYGDDLMKADIMSKIGGNSEWLTDVYKLGRQNEAPNLLGID 206
Query 175 ----------------VATAQRANLLLSSKAQEIMNMYLPQEKQIELSTLGAQYWNLIRD 218
VA AQ A L L ++ Q I+N ++P ++Q E A + +
Sbjct 207 LRKKRLENLSTETNIKVALAQGALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFAQYKA 266
Query 219 GSIKEEQAKNLLATRLEIAARTAGQHISNKVARSTADSIIDATNTAKMNEAAYNRGY--- 275
G + E Q K + + + A+ AGQ ++N++A AD A AAY G+
Sbjct 267 GKLSEAQVKTQIKQQALLEAQAAGQKLNNRLAERLADYQFKAMAAEYRANAAYYNGFYND 326
Query 276 -----------SQFSNDVGFRTGKMDRWLQDPVKARWDRG--INNAGEFIEGLSNIVGSV 322
++ ++ +M +D KA W N E ++GL VGSV
Sbjct 327 AWQAGMSKAAQVRYESNAARIAAQMSEIFKDREKASWKNNPIYYNISELLKGLLGSVGSV 386
Query 323 -------TKFGFLRNANQ 333
+K G L+ A +
Sbjct 387 VGPFSIASKIGSLKKAGK 404
> Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.protein.BACEGG.02723
Length=418
Score = 80.5 bits (197), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 96/375 (26%), Positives = 160/375 (43%), Gaps = 59/375 (16%)
Query 22 NNRQGSKNRKHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEW-----------NSAKN 70
+N+ ++ K E+ +I NE+ +SE+ K+RDF S +A+ +W N
Sbjct 27 SNKSVKRSIKAAKEINQINNEFNASEALKNRDFQTSEREASQQWNLDQWNRENAYNDPSA 86
Query 71 QRARLEEAGLNPYLM-MNggsagtasstsantvsgasgsggtPYQYTPTNIIGDVASFAG 129
QRAR+E AG NPY M ++ GSA T+ + S+ + + TP T D + A
Sbjct 87 QRARMEAAGFNPYNMNIDAGSASTSGAQSSPGSGSQATASHTPSLPAYTGYAADFQNVAS 146
Query 130 AMKSLSEARKTNTESDLLGKYGD----SDYSSRI-ANTE--ADTYFKQRQSD-------- 174
+ + A + ++ L YGD +D S+I N+E D Y RQ++
Sbjct 147 GIAQIGNAVSSGIDARLTSAYGDDLMKADIMSKIGGNSEWLTDVYKLGRQNEAPNLLGID 206
Query 175 ----------------VATAQRANLLLSSKAQEIMNMYLPQEKQIELSTLGAQYWNLIRD 218
VA AQ A L L ++ Q I+N ++P ++Q E A + +
Sbjct 207 LRKKRLENLSTETNIKVALAQGALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFAQYKA 266
Query 219 GSIKEEQAKNLLATRLEIAARTAGQHISNKVARSTADSIIDATNTAKMNEAAYNRGY--- 275
G + E Q K + + + A+T GQ ++N++A AD A AAY G+
Sbjct 267 GKLSEAQVKTQIKQQALLEAQTVGQKLNNRLAERLADYQFKAMAAEYRANAAYYNGFYND 326
Query 276 -----------SQFSNDVGFRTGKMDRWLQDPVKARWDRG--INNAGEFIEGLSNIVGSV 322
+++ ++ M + K+ W N GE ++G+ VGSV
Sbjct 327 AWQAGMSKAAQARYESNAARIAADMSEIFKGREKSSWKNNPIYYNIGELLKGILGSVGSV 386
Query 323 TKFGFLRNANQRLKE 337
+L + LK+
Sbjct 387 AGPLYLGSKVSSLKK 401
> Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.protein.BACEGG.02723
Length=427
Score = 77.4 bits (189), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 90/337 (27%), Positives = 156/337 (46%), Gaps = 53/337 (16%)
Query 34 LEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNPYLMMNggsagt 93
L++ ++ NE+ + E++K+R F M++ N +N+ QRARLEE G N Y+ A
Sbjct 63 LKINQMNNEFNAKEAEKARAFQLDMWNKENAYNTPAAQRARLEEGGYNAYM----NPADA 118
Query 94 asstsantvsgasgsggtPYQYTPTNIIGDV-ASFAGAMKSLSEARKTNTESDLLGKY-- 150
S++ ++ S AS + Q T + +G+V A +K+ SE + + + L Y
Sbjct 119 GSASGMSSTSAASAASSAVMQGTDFSSLGEVGVRLAQELKTFSEKKGLDIRNFSLKDYLQ 178
Query 151 -------GDSDYSSRIANTEADTY---------------FKQR------QSDVATAQRAN 182
GD+++ R + EA Y +++ +++ A AN
Sbjct 179 SQIDKMKGDTNW--RNVSPEAIRYNIMSGLEAAKIGMENLREQWANQVWSNNLLRANVAN 236
Query 183 LLLSSKAQEIMNMYLPQEKQIELSTLGAQYWNLIRDGSIKEEQAKNLLATRLEIAARTAG 242
LL ++++ I+N YL Q++Q +L+ A Y LI G + +A+ LL+ + AR G
Sbjct 237 SLLDAESKTILNKYLDQQQQADLNVKAAHYEELINRGQLHVVEARELLSREVLNYARARG 296
Query 243 QHISNKVARSTADSIIDATNTAKMNEAAYN---RGYSQFSNDVGFR--------TGKM-- 289
+ISN VA +A ++ A N A E +YN R Y + F TG++
Sbjct 297 LNISNWVAAKSAKGLVYANNAANYYEGSYNNYRRQYVKLDAASDFMGNQWQNQLTGELLN 356
Query 290 -DRWLQDPVKARWDRGINNAGEFIEGLSNIVGSVTKF 325
RW D W IN+A + G S+++ + +
Sbjct 357 SKRW--DNEMQAWREAINSANALLRGSSDVMDAYNTY 391
> Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2
Length=380
Score = 76.6 bits (187), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 83/289 (29%), Positives = 136/289 (47%), Gaps = 46/289 (16%)
Query 20 IGNNRQGSKNRKHQ----LEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARL 75
+ +N G N+ HQ L M R QN + + ++Q RD+ K M+ N +NS + +R
Sbjct 14 LTDNVVGMVNQNHQNKVNLRMMREQNAFNAEQAQIQRDWQKQMWGMNNAYNSPSSMISR- 72
Query 76 EEAGLNPYLMMNggsa-gtasstsantvsgasgsggtPYQYTPTNIIGDVASFA------ 128
GLNP++ + A + ++ + A Y+ +++ +AS A
Sbjct 73 ---GLNPFVQGSAAMAGSKSPASGGAAATAAPVPSMQAYKPNFSSVFQSLASLAQAKASE 129
Query 129 -GAMKSLSEARKTNTESDLLGKY------------GDSDY----SSRIA-----NTEA-- 164
A +S S AR+T+T + LL Y G S Y + R++ +TE
Sbjct 130 ASAGESGSRARQTDTVTPLLSDYYRGLTNWKNLAIGSSGYWNKETGRVSAALDQSTETQN 189
Query 165 --DTYFKQRQSDVATAQRANLLLSSKAQEIMNMYLPQEKQIELSTLGAQYWNLIRDGSIK 222
+ F +R ++ AQ A +LL+S AQ IMN Y+ Q +Q +L NL G++
Sbjct 190 LKNAQFAER---ISAAQEAQILLNSDAQRIMNKYMDQNQQADLFIKAQTLANLQSQGALT 246
Query 223 EEQAKNLLATRLEIAARTAGQHISNKVARSTADSIIDATNTAKMNEAAY 271
E+Q + + + +A +G+ I N+VA TADS+I A N + NE Y
Sbjct 247 EKQIQTEIQRAILASAEASGKKIDNRVASETADSLIKAANAS--NELQY 293
> Alpavirinae_Human_feces_A_021_Microviridae_AG079_hypothetical.protein.BACEGG.02723
Length=397
Score = 75.5 bits (184), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 36/62 (58%), Positives = 46/62 (74%), Gaps = 0/62 (0%)
Query 26 GSKNRKHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNPYLM 85
G K RK Q + + + S+E+QK+RDF M++ TNE+NSA NQRARLEEAGLNPY+M
Sbjct 2 GRKERKFQAQQAELNRNFQSAEAQKNRDFEVDMWNKTNEYNSATNQRARLEEAGLNPYMM 61
Query 86 MN 87
MN
Sbjct 62 MN 63
Score = 32.0 bits (71), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 30/127 (24%), Positives = 62/127 (49%), Gaps = 14/127 (11%)
Query 141 NTESDLLGKYGD-------SDYSSRIANTEADTYFKQRQSDVATAQRANLLLSSKAQEIM 193
++ S + GKYG+ + + + +++ D K + + A + LSS AQ+++
Sbjct 159 DSASRIFGKYGNWKQAGSAVMFDNAVQSSDLDRQNKNLTNQMIQANMIQVNLSSDAQKVI 218
Query 194 NMYLPQEKQIELSTLGAQYWNLIRDGSIKEEQAKNLL----ATRLEIAARTAGQHISNKV 249
N YL Q++ ++L+ A Y G + EQ + L + RL+ R A I+++
Sbjct 219 NKYLDQQESVKLNIQSALYTEAAARGQLTFEQWQGQLIQNMSDRLDYNTRKA---IADEF 275
Query 250 ARSTADS 256
R++ ++
Sbjct 276 IRASCEA 282
Lambda K H a alpha
0.313 0.127 0.359 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 31621345