bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
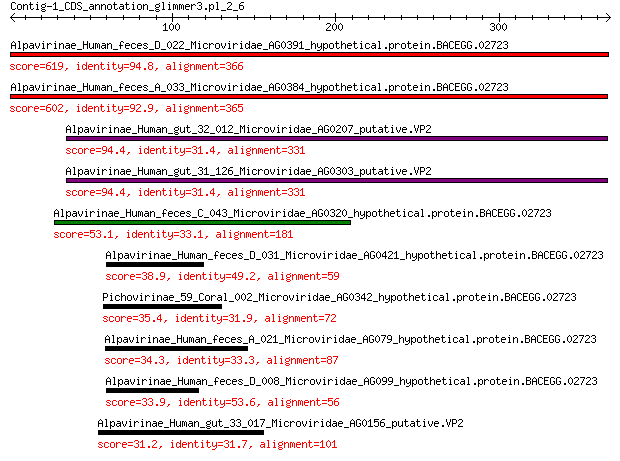
Query= Contig-1_CDS_annotation_glimmer3.pl_2_6
Length=367
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_D_022_Microviridae_AG0391_hypothetical.... 619 0.0
Alpavirinae_Human_feces_A_033_Microviridae_AG0384_hypothetical.... 602 0.0
Alpavirinae_Human_gut_32_012_Microviridae_AG0207_putative.VP2 94.4 2e-23
Alpavirinae_Human_gut_31_126_Microviridae_AG0303_putative.VP2 94.4 2e-23
Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.... 53.1 1e-09
Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.... 38.9 4e-05
Pichovirinae_59_Coral_002_Microviridae_AG0342_hypothetical.prot... 35.4 4e-04
Alpavirinae_Human_feces_A_021_Microviridae_AG079_hypothetical.p... 34.3 0.001
Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.p... 33.9 0.002
Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2 31.2 0.011
> Alpavirinae_Human_feces_D_022_Microviridae_AG0391_hypothetical.protein.BACEGG.02723
Length=367
Score = 619 bits (1595), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 347/366 (95%), Positives = 358/366 (98%), Gaps = 0/366 (0%)
Query 1 MSALVTsaiiagasglaaaggssiaasKMNSRAEKYNRWALKEQQRYQKEYADYMAQLEA 60
MSALVTSA+IAGAS LAAAGGSSIAASKMN+RAEKYNRWALKEQQRYQKEYADYMAQLE+
Sbjct 1 MSALVTSALIAGASSLAAAGGSSIAASKMNARAEKYNRWALKEQQRYQKEYADYMAQLES 60
Query 61 QQNNLYWDKYNSPAAQRRARVAAGFTPYADVGGIQTSSVDPGSYGGSTPSAQSFTQPGGI 120
QQNNLYWDKYNSPAAQRRARVAAG TPYADVGGIQTSSVDPGSYGGSTPSAQSFTQPGGI
Sbjct 61 QQNNLYWDKYNSPAAQRRARVAAGLTPYADVGGIQTSSVDPGSYGGSTPSAQSFTQPGGI 120
Query 121 PSSPLVGAFGNATQQTLSALQAEANIELTKSQALKTRAETTGLENTNSMFDVVKSIADEE 180
P+SPLVGAFGNATQQTLSALQAEANIELTKSQALKTRAETTGLENTNSMFD+VKSIA E+
Sbjct 121 PTSPLVGAFGNATQQTLSALQAEANIELTKSQALKTRAETTGLENTNSMFDIVKSIASED 180
Query 181 LTSKRFSNILKEVEVKYAEVNAITDLDAKQAKIAEINASALERLASAAKTDADRITVELL 240
LTSK+F+NILKE+E KYAE NAI DLD KQAKI EINASALERLASAAKTDADRITVELL
Sbjct 181 LTSKQFNNILKEIETKYAEANAIADLDTKQAKIGEINASALERLASAAKTDADRITVELL 240
Query 241 RDAQKRsleagaslaeaqaaTEPHRALNLKQDTLLKMAQEETEQLLRSQKFELTRQQARA 300
RDAQKRSLEAGASLAEAQAATEPHRALNLKQDTLLKMAQEETEQLLRSQKFELTRQQARA
Sbjct 241 RDAQKRSLEAGASLAEAQAATEPHRALNLKQDTLLKMAQEETEQLLRSQKFELTRQQARA 300
Query 301 AAISFVQERILTYRQAEELARYLANIHDPKNMWDGIWRIVSLPAGISKSDFAADLYNALY 360
AA+SFVQERILTYRQAEELARYLANIHDPKNMWDGIWR+VSLPAGISKSDFAADLYNALY
Sbjct 301 AAMSFVQERILTYRQAEELARYLANIHDPKNMWDGIWRLVSLPAGISKSDFAADLYNALY 360
Query 361 EEINSI 366
EEINSI
Sbjct 361 EEINSI 366
> Alpavirinae_Human_feces_A_033_Microviridae_AG0384_hypothetical.protein.BACEGG.02723
Length=367
Score = 602 bits (1551), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 339/365 (93%), Positives = 351/365 (96%), Gaps = 0/365 (0%)
Query 1 MSALVTsaiiagasglaaaggssiaasKMNSRAEKYNRWALKEQQRYQKEYADYMAQLEA 60
MSALVTSAIIAGAS LAAAGGS+ AASKMN+RAEKYNRWALKEQQRYQKEYADY+AQ+E
Sbjct 1 MSALVTSAIIAGASSLAAAGGSAAAASKMNARAEKYNRWALKEQQRYQKEYADYLAQIET 60
Query 61 QQNNLYWDKYNSPAAQRRARVAAGFTPYADVGGIQTSSVDPGSYGGSTPSAQSFTQPGGI 120
QQNNLYW+KYNSPAAQRRARVAAG TPYADVGGIQTSSVDPGSYGGSTPSAQSF+QPGGI
Sbjct 61 QQNNLYWEKYNSPAAQRRARVAAGLTPYADVGGIQTSSVDPGSYGGSTPSAQSFSQPGGI 120
Query 121 PSSPLVGAFGNATQQTLSALQAEANIELTKSQALKTRAETTGLENTNSMFDVVKSIADEE 180
P +PLVGAFGNATQQTLSALQAEANIELTKSQALKTRAETTGLENTNSMFD+VKSIA E+
Sbjct 121 PINPLVGAFGNATQQTLSALQAEANIELTKSQALKTRAETTGLENTNSMFDIVKSIARED 180
Query 181 LTSKRFSNILKEVEVKYAEVNAITDLDAKQAKIAEINASALERLASAAKTDADRITVELL 240
LTSKRF NILKEVE +YAE NAI DLD KQAKIAEINAS LERLASAAKTDADRITVELL
Sbjct 181 LTSKRFGNILKEVESRYAEANAIADLDTKQAKIAEINASVLERLASAAKTDADRITVELL 240
Query 241 RDAQKRsleagaslaeaqaaTEPHRALNLKQDTLLKMAQEETEQLLRSQKFELTRQQARA 300
RDAQKRSLEAGASLAEAQAATEPH+ALNLKQDTLLKMAQEETEQLLRSQKFELTRQQAR
Sbjct 241 RDAQKRSLEAGASLAEAQAATEPHKALNLKQDTLLKMAQEETEQLLRSQKFELTRQQARL 300
Query 301 AAISFVQERILTYRQAEELARYLANIHDPKNMWDGIWRIVSLPAGISKSDFAADLYNALY 360
AA+SFVQERILTYRQAEELARYLANIHDPKNMWDGIWRIVSLPAGISKSDFAADLYNALY
Sbjct 301 AAMSFVQERILTYRQAEELARYLANIHDPKNMWDGIWRIVSLPAGISKSDFAADLYNALY 360
Query 361 EEINS 365
EEI S
Sbjct 361 EEITS 365
> Alpavirinae_Human_gut_32_012_Microviridae_AG0207_putative.VP2
Length=364
Score = 94.4 bits (233), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 104/345 (30%), Positives = 163/345 (47%), Gaps = 27/345 (8%)
Query 35 KYNRWALKEQQRYQKEYADYMAQLEAQ---QNNLY---WDKYNSPAAQRRARVAAGFTPY 88
K +W +++ Y + AD A EAQ Q +L + ++SPAAQR A AAG P+
Sbjct 30 KNRKW---QEKMYNMQVADRRADAEAQAQRQKDLAQWAYTNFDSPAAQRAAYAAAGVNPF 86
Query 89 ADVGGIQTSSVDPGS----YGGSTPSAQSFTQPGGIPSSPLVGAFGNATQQTLSALQ--- 141
+ IQ + V+ GS GG P + + P S L + Q + Q
Sbjct 87 VEGSSIQPAQVNSGSAQAAQGGDVPGSGPYQMN---PMSALQSGASSIIQNAMLDRQMKN 143
Query 142 AEANIELTKSQALKTRAETTGLENTNSMFDVVKSIADEELTSKRFSNILKEVEVKYAEVN 201
A+A+I L +Q +KT AE G N NS+F+ +S A+ + SKRF L +V ++A+
Sbjct 144 ADADIALKDAQRIKTLAEAKGQTNENSLFEFARSAAESDALSKRFRAELDKVNAEFAQAF 203
Query 202 AITDLDAKQAKIAEINASALERLASAAKTDADRITVELLRDAQKRsleagaslaeaqaaT 261
A D+ ++A + EI + LA AA+TDADR+T++ +RD Q + +L + T
Sbjct 204 AEADISQRKALLQEIWSRVKNNLAQAARTDADRLTIDTIRDLQAK------ALGAGISLT 257
Query 262 EPHRALNLKQDTLLKMAQEETEQLLRSQKFELTRQQARAAAISFVQERILTYRQAEELAR 321
E + Q T L AQ ETE LR + LT+ Q R++T E L
Sbjct 258 ESQTSATQAQ-TGLTQAQTETENQLRKLRKALTQNQINEITQKIRSSRVVTAEGIERLTA 316
Query 322 YLANIHDPKNMWDGIWR-IVSLPAGISKSDFAADLYNALYEEINS 365
+L + +++ I + I + S + D+ LY+ +N
Sbjct 317 WLRGDREAGSLFGLIDKYITGTGKDLLSSKYNNDIRGYLYDLMNG 361
> Alpavirinae_Human_gut_31_126_Microviridae_AG0303_putative.VP2
Length=364
Score = 94.4 bits (233), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 104/345 (30%), Positives = 163/345 (47%), Gaps = 27/345 (8%)
Query 35 KYNRWALKEQQRYQKEYADYMAQLEAQ---QNNLY---WDKYNSPAAQRRARVAAGFTPY 88
K +W +++ Y + AD A EAQ Q +L + ++SPAAQR A AAG P+
Sbjct 30 KNRKW---QEKMYNMQVADRRADAEAQAQRQKDLAQWAYTNFDSPAAQRAAYAAAGVNPF 86
Query 89 ADVGGIQTSSVDPGS----YGGSTPSAQSFTQPGGIPSSPLVGAFGNATQQTLSALQ--- 141
+ IQ + V+ GS GG P + + P S L + Q + Q
Sbjct 87 VEGSSIQPAQVNSGSAQAAQGGDVPGSGPYQMN---PMSALQSGASSIIQNAMLDRQMKN 143
Query 142 AEANIELTKSQALKTRAETTGLENTNSMFDVVKSIADEELTSKRFSNILKEVEVKYAEVN 201
A+A+I L +Q +KT AE G N NS+F+ +S A+ + SKRF L +V ++A+
Sbjct 144 ADADIALKDAQRIKTLAEAKGQTNENSLFEFARSAAESDALSKRFRAELDKVNAEFAQAF 203
Query 202 AITDLDAKQAKIAEINASALERLASAAKTDADRITVELLRDAQKRsleagaslaeaqaaT 261
A D+ ++A + EI + LA AA+TDADR+T++ +RD Q + +L + T
Sbjct 204 AEADISQRKALLQEIWSRVKNNLAQAARTDADRLTIDTIRDLQAK------ALGAGISLT 257
Query 262 EPHRALNLKQDTLLKMAQEETEQLLRSQKFELTRQQARAAAISFVQERILTYRQAEELAR 321
E + Q T L AQ ETE LR + LT+ Q R++T E L
Sbjct 258 ESQTSATQAQ-TGLTQAQTETENQLRKLRKALTQNQINEITQKIRSSRVVTAEGIERLTA 316
Query 322 YLANIHDPKNMWDGIWR-IVSLPAGISKSDFAADLYNALYEEINS 365
+L + +++ I + I + S + D+ LY+ +N
Sbjct 317 WLRGDREAGSLFGLIDKYITGTGKDLLSSKYNNDIRGYLYDLMNG 361
> Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.protein.BACEGG.02723
Length=396
Score = 53.1 bits (126), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 60/198 (30%), Positives = 92/198 (46%), Gaps = 32/198 (16%)
Query 28 KMNSRAEKYNRWALKEQQRYQKEYA----DYMAQLEAQQN--NL-YWDK---YNSPAAQR 77
KMN RAEKYN+WAL++Q + ++ A D+ ++ +QQN NL W++ YN+PAAQR
Sbjct 29 KMNRRAEKYNKWALQQQMAFNEQQAQLGRDWSEEMMSQQNEWNLQQWNRENAYNTPAAQR 88
Query 78 RARVAAGFTPYADVGGIQTSSVDPGSYGGSTPSAQSFTQPGGIPSSPLVGAFGNATQQTL 137
AAG + G GS G S Q P G P + G+ +
Sbjct 89 SRLEAAGLNAALAMQG-------QGSI-GMAGSGQPAAAPAGSPQAATGGSSAPQYSRPD 140
Query 138 SALQAEANIELTKSQALKTRAETTGLENTNSMFDVVKSIADEELT-------SKRFSNIL 190
+L ++A K++ L ++ GL+N ++K+ +EL S SN+
Sbjct 141 FSLLSQAVDSFFKNKLLSEQSTGQGLDN------LLKARYGDELAQISIGKGSAEISNLR 194
Query 191 KEVEVKYAEVNAITDLDA 208
+ YAE A+ L A
Sbjct 195 SQSARNYAET-AVASLTA 211
> Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.protein.BACEGG.02723
Length=418
Score = 38.9 bits (89), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 29/63 (46%), Positives = 35/63 (56%), Gaps = 15/63 (24%)
Query 60 AQQNNL-YWDK---YNSPAAQRRARVAAGFTPYADVGGIQTSSVDPGSYGGSTPSAQSFT 115
+QQ NL W++ YN P+AQR AAGF PY ++DPGS GST AQS
Sbjct 67 SQQWNLDQWNRENAYNDPSAQRARMEAAGFNPY-------NMNIDPGS--GSTSGAQS-- 115
Query 116 QPG 118
PG
Sbjct 116 SPG 118
> Pichovirinae_59_Coral_002_Microviridae_AG0342_hypothetical.protein.BACEGG.02723
Length=233
Score = 35.4 bits (80), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/75 (31%), Positives = 35/75 (47%), Gaps = 5/75 (7%)
Query 58 LEAQQNNLYWD---KYNSPAAQRRARVAAGFTPYADVGGIQTSSVDPGSYGGSTPSAQSF 114
L +QN +W+ KYN+P AQ AG P G QT++ GS S P+ +
Sbjct 29 LANRQNVEFWNMQNKYNTPKAQMERLKEAGLNPNLIYGSGQTNTGVAGSIAASKPAPYNI 88
Query 115 TQPGGIPSSPLVGAF 129
P +P++ G +
Sbjct 89 QNP--VPAAIAAGMY 101
> Alpavirinae_Human_feces_A_021_Microviridae_AG079_hypothetical.protein.BACEGG.02723
Length=397
Score = 34.3 bits (77), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/94 (31%), Positives = 40/94 (43%), Gaps = 20/94 (21%)
Query 59 EAQQNNLY----WDK---YNSPAAQRRARVAAGFTPYADVGGIQTSSVDPGSYGGSTPSA 111
EAQ+N + W+K YNS QR AG PY + GG+ A
Sbjct 23 EAQKNRDFEVDMWNKTNEYNSATNQRARLEEAGLNPYMMMN------------GGNAGEA 70
Query 112 QSFTQPGGIPSSPLVGAFGNATQQTLSALQAEAN 145
S T P P + GA G+ T+ +S L + +N
Sbjct 71 GSVTAP-STPQGAMPGATGDTTENVISGLNSVSN 103
> Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.protein.BACEGG.02723
Length=418
Score = 33.9 bits (76), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 36/71 (51%), Gaps = 17/71 (24%)
Query 60 AQQNNL-YWDK---YNSPAAQRRARVAAGFTPY---ADVGGIQTSSV--DPGSYGGS--- 107
+QQ NL W++ YN P+AQR AAGF PY D G TS PGS GS
Sbjct 67 SQQWNLDQWNRENAYNDPSAQRARMEAAGFNPYNMNIDAGSASTSGAQSSPGS--GSQAT 124
Query 108 ---TPSAQSFT 115
TPS ++T
Sbjct 125 ASHTPSLPAYT 135
> Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2
Length=353
Score = 31.2 bits (69), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 51/110 (46%), Gaps = 13/110 (12%)
Query 55 MAQLEAQQNNLYWDK---YNSPAAQRRARVAAGFTPYADV-GGIQTSSVDPGSYGGSTPS 110
M Q + Q+N W+K YN+ +AQR+ AG PY + GG S G+ ++ S
Sbjct 51 MQQRDFQEN--MWNKENTYNTASAQRQRLEEAGLNPYLMMNGGSSGVSQSAGTGASASSS 108
Query 111 AQSFTQPGGIPSSPLVGAFGNATQQ-----TLSALQAEANIELTKSQALK 155
+ QP S + A G+ Q +S +Q + N L +QA++
Sbjct 109 GTAVFQPFQADFSGIQQAIGSVFQSQVRQAQVSQMQGQRN--LADAQAMQ 156
Lambda K H a alpha
0.312 0.125 0.339 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 31835727