bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-23_CDS_annotation_glimmer3.pl_2_5
Length=294
Score E
Sequences producing significant alignments: (Bits) Value
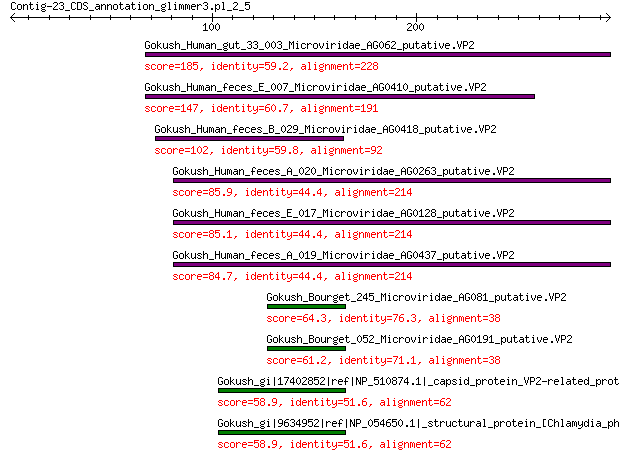
Gokush_Human_gut_33_003_Microviridae_AG062_putative.VP2 185 7e-58
Gokush_Human_feces_E_007_Microviridae_AG0410_putative.VP2 147 2e-43
Gokush_Human_feces_B_029_Microviridae_AG0418_putative.VP2 102 4e-27
Gokush_Human_feces_A_020_Microviridae_AG0263_putative.VP2 85.9 2e-21
Gokush_Human_feces_E_017_Microviridae_AG0128_putative.VP2 85.1 4e-21
Gokush_Human_feces_A_019_Microviridae_AG0437_putative.VP2 84.7 7e-21
Gokush_Bourget_245_Microviridae_AG081_putative.VP2 64.3 6e-14
Gokush_Bourget_052_Microviridae_AG0191_putative.VP2 61.2 9e-13
Gokush_gi|17402852|ref|NP_510874.1|_capsid_protein_VP2-related_... 58.9 2e-12
Gokush_gi|9634952|ref|NP_054650.1|_structural_protein_[Chlamydi... 58.9 2e-12
> Gokush_Human_gut_33_003_Microviridae_AG062_putative.VP2
Length=295
Score = 185 bits (470), Expect = 7e-58, Method: Compositional matrix adjust.
Identities = 135/230 (59%), Positives = 162/230 (70%), Gaps = 6/230 (3%)
Query 67 TNDDQIAKYLQQAYGYQSAEGIAQQKYNQKSMLQQMGYNTMQAITQGIYNHIENTAAMNY 126
TND+QI KYL + Y +Q + Q K N+++ML QMGYNT+ AI QGIYNHIE AAMNY
Sbjct 70 TNDEQIMKYLDRFYAWQGGQNAFQSKTNRQNMLMQMGYNTLGAIQQGIYNHIEQNAAMNY 129
Query 127 NSAEALANREFQERMSNTAYQRTVEDMKKAGLNPILAFanggastpggstatisgasmgl 186
NSAEALANR FQERMS+T+YQR VEDM+KAGLNPILAFANGGASTPGGS ATI+GASMG+
Sbjct 130 NSAEALANRNFQERMSSTSYQRAVEDMRKAGLNPILAFANGGASTPGGSGATITGASMGM 189
Query 187 asssalgisrsNGFVPNsysstswsksDWYNASQSWNQMLSETHLSPYGLQKALTNIGNK 246
SSSALG+S NG VP S S S S + WY +++ LS +H +P KAL + K
Sbjct 190 PSSSALGVSTMNGNVPTSNYSRSESNAQWYQLAEAVGSQLSTSHSTP----KALVDDLLK 245
Query 247 TDETIEKVTNKMGKKAAGSNERGHKPNNPM--QDKTGSYGQKRKPGDYLK 294
T + +EK + A G K + + QDKTG YG+KRKPGDYLK
Sbjct 246 TYKAMEKTEETVPGAAGGGGRSKTKQDRAIKPQDKTGKYGEKRKPGDYLK 295
> Gokush_Human_feces_E_007_Microviridae_AG0410_putative.VP2
Length=270
Score = 147 bits (370), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 116/191 (61%), Positives = 141/191 (74%), Gaps = 5/191 (3%)
Query 67 TNDDQIAKYLQQAYGYQSAEGIAQQKYNQKSMLQQMGYNTMQAITQGIYNHIENTAAMNY 126
TND QI YL + Y +Q + Q K N+++ML QMGYNT+ AI QGIYNHIEN AAM Y
Sbjct 67 TNDKQIMDYLNRYYQWQGGQNAFQSKTNRQNMLMQMGYNTLSAIQQGIYNHIENNAAMQY 126
Query 127 NSAEALANREFQERMSNTAYQRTVEDMKKAGLNPILAFanggastpggstatisgasmgl 186
NSAEALANR+FQERMS+TAYQR VEDM+KAGLNPILA+A GGASTPGGS ATI+GASMG+
Sbjct 127 NSAEALANRQFQERMSSTAYQRAVEDMRKAGLNPILAYAQGGASTPGGSGATITGASMGM 186
Query 187 asssalgisrsNGFVPNsysstswsksDWYNASQSWNQMLSETHLSPYGLQKALTNIGNK 246
+SSALG+S +G VPNSY + S SKS WY +++ +S + SP L + L K
Sbjct 187 PTSSALGVSTLSGNVPNSYFNRSESKSQWYQLAEAVGSQMSTGYSSPVQLTEDLL----K 242
Query 247 TDETIEKVTNK 257
T + +EK TNK
Sbjct 243 TYKQMEK-TNK 252
> Gokush_Human_feces_B_029_Microviridae_AG0418_putative.VP2
Length=272
Score = 102 bits (254), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 55/96 (57%), Positives = 67/96 (70%), Gaps = 11/96 (11%)
Query 72 IAKYL----QQAYGYQSAEGIAQQKYNQKSMLQQMGYNTMQAITQGIYNHIENTAAMNYN 127
+AKY QQA G QS + N K+ L +G NT+ AI QG+YN I+ AAM+YN
Sbjct 74 LAKYFLGQSQQAQGMQSLQ-------NNKNSLMALGLNTLGAIQQGVYNRIQQDAAMSYN 126
Query 128 SAEALANREFQERMSNTAYQRTVEDMKKAGLNPILA 163
SAEA ANR +QERMSNT+YQR EDM+KAG+NPILA
Sbjct 127 SAEAAANRAWQERMSNTSYQRATEDMRKAGINPILA 162
> Gokush_Human_feces_A_020_Microviridae_AG0263_putative.VP2
Length=300
Score = 85.9 bits (211), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 95/216 (44%), Positives = 118/216 (55%), Gaps = 12/216 (6%)
Query 81 GYQSAEGIAQQKYNQKSMLQQMGYNTMQAITQGIYNHIENTAAMNYNSAEALANREFQER 140
G SA+GI YN Q N+M QG N AM YNSAEA NRE+QE
Sbjct 95 GSMSAQGI----YNMAGSGFQGLLNSMMMNKQGSMNAALMREAMAYNSAEAALNREWQEH 150
Query 141 MSNTAYQRTVEDMKKAGLNPILAFanggastpggstatisgasmglasssalgisrsNGF 200
MS+TAYQR V DM+ AG+NPILA NGGA+ GGS ++ GAS+GL S+SA IS
Sbjct 151 MSSTAYQRAVADMRAAGINPILAALNGGAAMGGGSAGSVGGASVGLGSTSAASISALG-- 208
Query 201 VPNsysstswsksDWYNASQSWNQMLSETHLSPYGLQKALTNIGNKTDETIEKV--TNKM 258
P S++ S SD N +Q + + S Q+ N+ TD+ E K
Sbjct 209 SPAFSSNSYGSFSDSINLAQGVSSYFQQGANSGKSWQELKKNVEVVTDKVAEPSYDAGKA 268
Query 259 GKKAAGSNERGHKPNNPMQDKTGSYGQKRKPGDYLK 294
K+ AGS +R K DKTGSYGQ+RKPGDYL+
Sbjct 269 QKRVAGSTDRAIKAT----DKTGSYGQQRKPGDYLR 300
> Gokush_Human_feces_E_017_Microviridae_AG0128_putative.VP2
Length=300
Score = 85.1 bits (209), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 95/216 (44%), Positives = 118/216 (55%), Gaps = 12/216 (6%)
Query 81 GYQSAEGIAQQKYNQKSMLQQMGYNTMQAITQGIYNHIENTAAMNYNSAEALANREFQER 140
G SA+GI YN Q N+M QG N AM YNSAEA NRE+QE
Sbjct 95 GSMSAQGI----YNMAGSGFQGLLNSMMMNKQGRMNADLMREAMAYNSAEAALNREWQEH 150
Query 141 MSNTAYQRTVEDMKKAGLNPILAFanggastpggstatisgasmglasssalgisrsNGF 200
MS+TAYQR V DM+ AG+NPILA NGGA+ GGS ++ GAS+GL S+SA IS
Sbjct 151 MSSTAYQRAVADMRAAGINPILAALNGGAAMGGGSAGSVGGASVGLGSTSAASISALG-- 208
Query 201 VPNsysstswsksDWYNASQSWNQMLSETHLSPYGLQKALTNIGNKTDETIEKV--TNKM 258
P S++ S SD N +Q + + S Q+ N+ TD+ E K
Sbjct 209 SPAFSSNSYGSFSDSINLAQGVSSYFQQGANSGKSWQELKKNVEVVTDKVAEPSYDAGKA 268
Query 259 GKKAAGSNERGHKPNNPMQDKTGSYGQKRKPGDYLK 294
K+ AGS +R K DKTGSYGQ+RKPGDYL+
Sbjct 269 QKRVAGSTDRAIKAT----DKTGSYGQQRKPGDYLR 300
> Gokush_Human_feces_A_019_Microviridae_AG0437_putative.VP2
Length=300
Score = 84.7 bits (208), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 95/216 (44%), Positives = 119/216 (55%), Gaps = 12/216 (6%)
Query 81 GYQSAEGIAQQKYNQKSMLQQMGYNTMQAITQGIYNHIENTAAMNYNSAEALANREFQER 140
G SA+GI YN Q N+M QG N AM YNSAEA NRE+QE
Sbjct 95 GSMSAQGI----YNMAGSGFQGLLNSMMMNKQGRMNAELMREAMAYNSAEAAHNREWQEY 150
Query 141 MSNTAYQRTVEDMKKAGLNPILAFanggastpggstatisgasmglasssalgisrsNGF 200
MS+TAYQR V DM+ AG+NPILA NGGA+ GGS ++ GAS+GL S+SA IS G
Sbjct 151 MSSTAYQRAVADMRAAGINPILAALNGGAAMGGGSAGSVGGASVGLGSTSAASISALGG- 209
Query 201 VPNsysstswsksDWYNASQSWNQMLSETHLSPYGLQKALTNIGNKTDETIEKV--TNKM 258
P S++ S SD N +Q + + S Q+ N+ TD+ + K
Sbjct 210 -PAFSSNSYGSFSDSINLAQGVSSYFQQGANSGRSWQQLKNNVEVVTDKVAKPSYDAGKA 268
Query 259 GKKAAGSNERGHKPNNPMQDKTGSYGQKRKPGDYLK 294
K+ AGS +R K DKTGSYGQKR+PGDYL+
Sbjct 269 QKRVAGSTDRAIKAT----DKTGSYGQKRQPGDYLR 300
> Gokush_Bourget_245_Microviridae_AG081_putative.VP2
Length=242
Score = 64.3 bits (155), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 29/38 (76%), Positives = 33/38 (87%), Gaps = 0/38 (0%)
Query 127 NSAEALANREFQERMSNTAYQRTVEDMKKAGLNPILAF 164
N AE+ NREFQER+SNTAYQR VEDMK AGLNP+LA+
Sbjct 40 NMAESQRNREFQERLSNTAYQRQVEDMKSAGLNPMLAY 77
> Gokush_Bourget_052_Microviridae_AG0191_putative.VP2
Length=253
Score = 61.2 bits (147), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 27/38 (71%), Positives = 33/38 (87%), Gaps = 0/38 (0%)
Query 127 NSAEALANREFQERMSNTAYQRTVEDMKKAGLNPILAF 164
N+A+A NR+FQERMS+TAYQR V DMK AGLNP+LA+
Sbjct 42 NAAQAQHNRDFQERMSSTAYQRAVSDMKAAGLNPMLAY 79
> Gokush_gi|17402852|ref|NP_510874.1|_capsid_protein_VP2-related_protein_[Guinea_pig_Chlamydia_phage]
Length=186
Score = 58.9 bits (141), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 32/65 (49%), Positives = 40/65 (62%), Gaps = 6/65 (9%)
Query 103 GYNTMQAITQGIYNHIENTAAMNYNSAEALANRE---FQERMSNTAYQRTVEDMKKAGLN 159
G + + I G+ ++ A N+ RE FQERMSNTAYQR +EDMKKAGLN
Sbjct 20 GLSFLPGIASGVLGYL---GAQKQNATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLN 76
Query 160 PILAF 164
P+LAF
Sbjct 77 PMLAF 81
> Gokush_gi|9634952|ref|NP_054650.1|_structural_protein_[Chlamydia_phage_2]
Length=186
Score = 58.9 bits (141), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 32/65 (49%), Positives = 40/65 (62%), Gaps = 6/65 (9%)
Query 103 GYNTMQAITQGIYNHIENTAAMNYNSAEALANRE---FQERMSNTAYQRTVEDMKKAGLN 159
G + + I G+ ++ A N+ RE FQERMSNTAYQR +EDMKKAGLN
Sbjct 20 GLSFLPGIASGVLGYL---GAQKQNATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLN 76
Query 160 PILAF 164
P+LAF
Sbjct 77 PMLAF 81
Lambda K H a alpha
0.310 0.125 0.355 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 24485066