bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-25_CDS_annotation_glimmer3.pl_2_6
Length=163
Score E
Sequences producing significant alignments: (Bits) Value
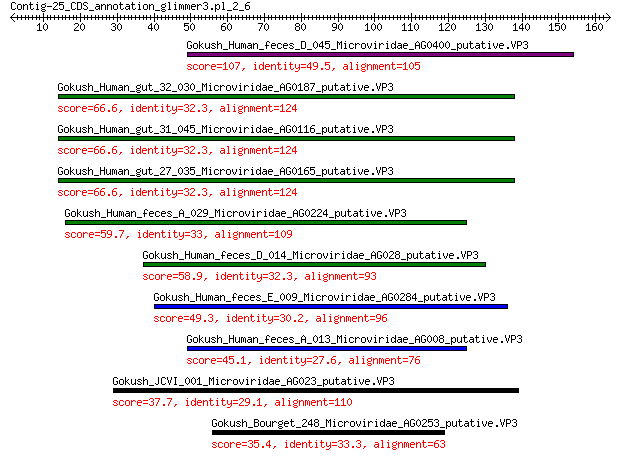
Gokush_Human_feces_D_045_Microviridae_AG0400_putative.VP3 107 1e-31
Gokush_Human_gut_32_030_Microviridae_AG0187_putative.VP3 66.6 4e-16
Gokush_Human_gut_31_045_Microviridae_AG0116_putative.VP3 66.6 4e-16
Gokush_Human_gut_27_035_Microviridae_AG0165_putative.VP3 66.6 4e-16
Gokush_Human_feces_A_029_Microviridae_AG0224_putative.VP3 59.7 1e-13
Gokush_Human_feces_D_014_Microviridae_AG028_putative.VP3 58.9 3e-13
Gokush_Human_feces_E_009_Microviridae_AG0284_putative.VP3 49.3 6e-10
Gokush_Human_feces_A_013_Microviridae_AG008_putative.VP3 45.1 2e-08
Gokush_JCVI_001_Microviridae_AG023_putative.VP3 37.7 9e-06
Gokush_Bourget_248_Microviridae_AG0253_putative.VP3 35.4 6e-05
> Gokush_Human_feces_D_045_Microviridae_AG0400_putative.VP3
Length=109
Score = 107 bits (267), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 52/107 (49%), Positives = 78/107 (73%), Gaps = 4/107 (4%)
Query 49 MEAEAPSTDINYMLHRLSLGDTSVLSSKRPMYGDFTGLPSDPIEALNLVHQSEYAFAQLS 108
+++ AP TD+NYMLHRLS+GDTSVLS ++ +YGDF+ LP +P++ +N+++ +E +F+QL
Sbjct 3 IDSFAPYTDLNYMLHRLSVGDTSVLSGRQAIYGDFSALPQNPVDMINVLNSAEQSFSQLP 62
Query 109 ADDKAKYNNDWRRWFADLLSGRDNLSEKLSSVVPNSDV--EKEGADS 153
AD+KA +NND+R W A+LL+ N +S VP+ EK G DS
Sbjct 63 ADEKAAFNNDYRVWLANLLNSASNPDVDIS--VPDKPAVEEKGGVDS 107
> Gokush_Human_gut_32_030_Microviridae_AG0187_putative.VP3
Length=157
Score = 66.6 bits (161), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 40/125 (32%), Positives = 63/125 (50%), Gaps = 3/125 (2%)
Query 14 IYTASGSPIHRILEPRFDGVN-TCLVVTGEENIQDRMEAEAPSTDINYMLHRLSLGDTSV 72
I++ GSP H +D L +G EN+ D +++ A S DI+ ++ R + GD
Sbjct 13 IFSDPGSPEHITYAGHYDEKGRVVLEESGRENLYDYIQSYAESCDIHVLMKRYANGDVDA 72
Query 73 LSSKRPMYGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRRWFADLLSGRDN 132
LS K+ YGDF P EALN +++ E F L + + K+ N + + A SG +
Sbjct 73 LSQKQGFYGDFLDFPKTYAEALNHMNEMERQFMALPVETREKFGNSFTEFLA--ASGEAD 130
Query 133 LSEKL 137
+KL
Sbjct 131 FFDKL 135
> Gokush_Human_gut_31_045_Microviridae_AG0116_putative.VP3
Length=157
Score = 66.6 bits (161), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 40/125 (32%), Positives = 63/125 (50%), Gaps = 3/125 (2%)
Query 14 IYTASGSPIHRILEPRFDGVN-TCLVVTGEENIQDRMEAEAPSTDINYMLHRLSLGDTSV 72
I++ GSP H +D L +G EN+ D +++ A S DI+ ++ R + GD
Sbjct 13 IFSDPGSPEHITYAGHYDEKGRVVLEESGRENLYDYIQSYAESCDIHVLMKRYANGDVDA 72
Query 73 LSSKRPMYGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRRWFADLLSGRDN 132
LS K+ YGDF P EALN +++ E F L + + K+ N + + A SG +
Sbjct 73 LSQKQGFYGDFLDFPKTYAEALNHMNEMERQFMALPVETREKFGNSFTEFLA--ASGEAD 130
Query 133 LSEKL 137
+KL
Sbjct 131 FFDKL 135
> Gokush_Human_gut_27_035_Microviridae_AG0165_putative.VP3
Length=157
Score = 66.6 bits (161), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 40/125 (32%), Positives = 63/125 (50%), Gaps = 3/125 (2%)
Query 14 IYTASGSPIHRILEPRFDGVN-TCLVVTGEENIQDRMEAEAPSTDINYMLHRLSLGDTSV 72
I++ GSP H +D L +G EN+ D +++ A S DI+ ++ R + GD
Sbjct 13 IFSDPGSPEHITYAGHYDEKGRVVLEESGRENLYDYIQSYAESCDIHVLMKRYANGDVDA 72
Query 73 LSSKRPMYGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRRWFADLLSGRDN 132
LS K+ YGDF P EALN +++ E F L + + K+ N + + A SG +
Sbjct 73 LSQKQGFYGDFLDFPKTYAEALNHMNEMERQFMALPVETREKFGNSFTEFLA--ASGEAD 130
Query 133 LSEKL 137
+KL
Sbjct 131 FFDKL 135
> Gokush_Human_feces_A_029_Microviridae_AG0224_putative.VP3
Length=162
Score = 59.7 bits (143), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 36/110 (33%), Positives = 56/110 (51%), Gaps = 1/110 (1%)
Query 16 TASGSPIHRILEPRFDGVNTC-LVVTGEENIQDRMEAEAPSTDINYMLHRLSLGDTSVLS 74
T GS E R++ L G+E+ +++ S DI+ +L R GD VL+
Sbjct 16 TGVGSREKITYEARYNAKGQLELNEKGKEDWYGYIQSHKDSVDIHVLLERFQRGDVDVLN 75
Query 75 SKRPMYGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRRWFA 124
+ YGD T PS +ALN+V SE F L +++AKYN+++ + A
Sbjct 76 RVQGFYGDITSYPSTFADALNIVRSSEEFFNSLPVEERAKYNHNFSEFLA 125
> Gokush_Human_feces_D_014_Microviridae_AG028_putative.VP3
Length=174
Score = 58.9 bits (141), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 49/93 (53%), Gaps = 0/93 (0%)
Query 37 LVVTGEENIQDRMEAEAPSTDINYMLHRLSLGDTSVLSSKRPMYGDFTGLPSDPIEALNL 96
L V G ENI ++++ T I ++ R GD + L + +Y D + +P++ IEA
Sbjct 58 LKVVGRENIYEKIQECLEPTKIENIIRRFEEGDPTALGHESGIYADISDMPTNIIEAQKR 117
Query 97 VHQSEYAFAQLSADDKAKYNNDWRRWFADLLSG 129
+ + FA L D K K+ ND + A++LSG
Sbjct 118 IQDVQAKFASLPIDIKEKFGNDPTVFMAEILSG 150
> Gokush_Human_feces_E_009_Microviridae_AG0284_putative.VP3
Length=148
Score = 49.3 bits (116), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 55/99 (56%), Gaps = 3/99 (3%)
Query 40 TGEENIQDRMEAEAPSTDINYMLHR-LSLGDTSVLSSKRPMYGDFTGLPSDPIEALNLVH 98
+G+ NI D++++ S DIN ++ R ++ GD S+LS + YGDF+ +P + LN +
Sbjct 32 SGKINIYDQIQSHKDSCDINLLIQRCVATGDESILSRVQGAYGDFSDMPHTYADMLNRLR 91
Query 99 QSEYAFAQLSADDKAKYNNDWRRWFA--DLLSGRDNLSE 135
++ F L + K++ ++ ++ + D DN SE
Sbjct 92 EAREFFDGLPLPTRQKFDCNFEQFISAMDKPGFLDNFSE 130
> Gokush_Human_feces_A_013_Microviridae_AG008_putative.VP3
Length=160
Score = 45.1 bits (105), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 21/76 (28%), Positives = 39/76 (51%), Gaps = 0/76 (0%)
Query 49 MEAEAPSTDINYMLHRLSLGDTSVLSSKRPMYGDFTGLPSDPIEALNLVHQSEYAFAQLS 108
+++ A S D++ ++ R + GD LS + YGD P EALN +++ E F L
Sbjct 48 IQSHAESVDLHVLMERYARGDVDALSKAQGFYGDVLDFPKTYAEALNHMNEMERQFMSLP 107
Query 109 ADDKAKYNNDWRRWFA 124
+ + K+ + + + A
Sbjct 108 VEIREKFGHSFTEFLA 123
> Gokush_JCVI_001_Microviridae_AG023_putative.VP3
Length=142
Score = 37.7 bits (86), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 50/110 (45%), Gaps = 4/110 (4%)
Query 29 RFDGVNTCLVVTGEENIQDRMEAEAPSTDINYMLHRLSLGDTSVLSSKRPMYGDFTGLPS 88
R + N V EE + + E DIN ++ + + S PM DF G+ +
Sbjct 11 RNEASNISAVAFPEETLAQQHFKE--QCDINRIVAQYAKTGLVPQSVATPMAEDFVGI-T 67
Query 89 DPIEALNLVHQSEYAFAQLSADDKAKYNNDWRRWFADLLSGRDNLSEKLS 138
D A+N + + + AF L+AD + K+ ND R F D NL E ++
Sbjct 68 DYHTAMNAIRRGDEAFNTLNADVREKFKNDAGR-FVDFCLDPANLDEVIA 116
> Gokush_Bourget_248_Microviridae_AG0253_putative.VP3
Length=145
Score = 35.4 bits (80), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 33/63 (52%), Gaps = 1/63 (2%)
Query 56 TDINYMLHRLSLGDTSVLSSKRPMYGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKY 115
DIN +L + ++ + P YGDFTG+ D A+N V + F L A +A++
Sbjct 41 CDINTILQKFNITGLLPEQTLSPRYGDFTGI-GDYHTAMNRVLAVQDEFEALPAQIRARF 99
Query 116 NND 118
+ND
Sbjct 100 DND 102
Lambda K H a alpha
0.311 0.130 0.371 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 11290891