bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-2_CDS_annotation_glimmer3.pl_2_1
Length=206
Score E
Sequences producing significant alignments: (Bits) Value
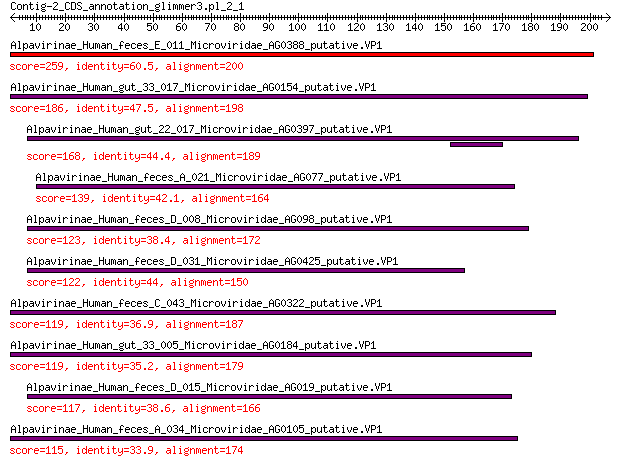
Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1 259 4e-84
Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1 186 1e-56
Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1 168 3e-50
Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1 139 5e-40
Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1 123 4e-34
Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1 122 1e-33
Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1 119 8e-33
Alpavirinae_Human_gut_33_005_Microviridae_AG0184_putative.VP1 119 2e-32
Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1 117 3e-32
Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1 115 2e-31
> Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1
Length=622
Score = 259 bits (662), Expect = 4e-84, Method: Compositional matrix adjust.
Identities = 121/200 (61%), Positives = 152/200 (76%), Gaps = 4/200 (2%)
Query 1 MAHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRP 60
MAHFTGLK+LQNHPH++GFD+G+KN+F+AK GEL+PVYWD IP C YDID+ YFTRTRP
Sbjct 1 MAHFTGLKELQNHPHKAGFDVGSKNLFTAKVGELIPVYWDFAIPDCDYDIDLAYFTRTRP 60
Query 61 VQTAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSL 120
VQTAAYTR+REYFDFYAVP DL+WKSFD++VIQMG+TAPVQ+K +L LTV D+P+C+L
Sbjct 61 VQTAAYTRVREYFDFYAVPCDLLWKSFDSAVIQMGQTAPVQSKTLLDPLTVGNDIPWCTL 120
Query 121 SDLGLSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNNMPAL 180
DL + +F+SGS + S S + + NIFGY RGDV+ KL+ LNYGN + N
Sbjct 121 LDLSNAVYFSSGSSPLGSTVS--VPSGFGNIFGYNRGDVDSKLLFYLNYGNFV--NPSLS 176
Query 181 NIGNSNYRWWNREAKKQDRP 200
N+G+ + RWWN P
Sbjct 177 NVGSPSNRWWNTSFSSSKVP 196
> Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1
Length=614
Score = 186 bits (471), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 94/198 (47%), Positives = 128/198 (65%), Gaps = 11/198 (6%)
Query 1 MAHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRP 60
MA +TGL LQNHPHRSGFDIG KN F+AK GELLPVYWD+ +PG Y +++YFTRT+P
Sbjct 1 MAFYTGLSNLQNHPHRSGFDIGRKNAFTAKVGELLPVYWDISMPGDKYRFNVEYFTRTQP 60
Query 61 VQTAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSL 120
V T+AYTR+REYFDFYAVP+ L+WKS + + QM + +QA + L++ LP ++
Sbjct 61 VATSAYTRLREYFDFYAVPLRLLWKSAPSVLTQMQDVNQIQALSLTQNLSLGTYLPSLTI 120
Query 121 SDLGLSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNNMPAL 180
LG + + +G+ P S+ N FG+ R D+++KL+ L YGN+I P+L
Sbjct 121 GTLGWAIRYLNGNTWEPETASY-----LRNAFGFSRADLSFKLLSYLGYGNLI-ETPPSL 174
Query 181 NIGNSNYRWWNREAKKQD 198
GN RWW+ K D
Sbjct 175 --GN---RWWSTSLKNTD 187
> Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1
Length=607
Score = 168 bits (425), Expect = 3e-50, Method: Compositional matrix adjust.
Identities = 84/189 (44%), Positives = 114/189 (60%), Gaps = 11/189 (6%)
Query 7 LKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAY 66
+ LQNHPHRSGFDIG KN F+AK GELLPVYWD+ +PG Y +++YFTRT+PV+T+AY
Sbjct 1 MSNLQNHPHRSGFDIGRKNAFTAKVGELLPVYWDISMPGDKYKFNVEYFTRTQPVETSAY 60
Query 67 TRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSLSDLGLS 126
TR+REYFDFYAVP+ L+WKS + + QM + +QA L++ P +LS
Sbjct 61 TRLREYFDFYAVPLRLLWKSAPSVLTQMQDINQLQALSFTQNLSLGSYFPSLTLSRFTAV 120
Query 127 CFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNNMPALNIGNSN 186
+G +VP S + N FG+ R D+ +KL L YGN+ + + N
Sbjct 121 LNRLNGGSNVPGNSS-----TFLNEFGFSRADLAFKLFSYLGYGNVWSSEFSSSN----- 170
Query 187 YRWWNREAK 195
RWW+ K
Sbjct 171 -RWWSTSLK 178
Score = 22.3 bits (46), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 10/18 (56%), Positives = 12/18 (67%), Gaps = 0/18 (0%)
Query 152 FGYIRGDVNYKLIHMLNY 169
FGY GDVN + +LNY
Sbjct 542 FGYSEGDVNSQNKVVLNY 559
> Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1
Length=622
Score = 139 bits (351), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 69/164 (42%), Positives = 103/164 (63%), Gaps = 7/164 (4%)
Query 10 LQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAYTRI 69
L+NHP RSGFD+ + F++K GELLPVYW IPG ++I +FTRT+PV TAAYTR+
Sbjct 9 LKNHPRRSGFDLTKRICFTSKAGELLPVYWKPTIPGDKWNISTNWFTRTQPVDTAAYTRV 68
Query 70 REYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSLSDLGLSCFF 129
+EY D++ VP++LI K ++++ QM + PV A + ++ D+PY +L L + +
Sbjct 69 KEYVDWFFVPLNLIQKGIESAITQMVDN-PVSAMSAIENRAITTDMPYTTLLSLSRALYM 127
Query 130 ASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNII 173
+G V S + N+FG+ R D++ KL+ ML YGN I
Sbjct 128 LNGKSYVNS------HAGKLNMFGFSRADLSAKLLQMLKYGNFI 165
> Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1
Length=618
Score = 123 bits (308), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 66/172 (38%), Positives = 101/172 (59%), Gaps = 4/172 (2%)
Query 7 LKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAY 66
+ ++NHP RSGFD+ + F++K GELLPV+WD+ PG ++ I Q FTRT+P+ TAAY
Sbjct 6 MSAVKNHPRRSGFDLSNRVCFTSKAGELLPVFWDIVYPGDSFKIKTQLFTRTQPLNTAAY 65
Query 67 TRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSLSDLGLS 126
TRIREY DFY VP+ LI K+ ++ QM + PVQA + + V+ D+P+ + S
Sbjct 66 TRIREYLDFYFVPLRLINKNLPTALTQMQDN-PVQATGLSSNKVVTTDIPWVPVHSSSGS 124
Query 127 CFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNNMP 178
+G V + +++ N G+ + KL+ L YGN + + +P
Sbjct 125 YSSLTGFADV---RGSVSSSDIENFLGFDSITQSAKLLMYLRYGNFLSSVVP 173
> Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1
Length=614
Score = 122 bits (305), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 66/160 (41%), Positives = 95/160 (59%), Gaps = 11/160 (7%)
Query 7 LKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAY 66
+ ++NHP RSGFD+ + F++K GELLPV+WD+ PG ++ I Q FTRT+P+ TAAY
Sbjct 1 MSAVKNHPRRSGFDLSNRVCFTSKAGELLPVFWDIVYPGDSFKIKTQLFTRTQPLNTAAY 60
Query 67 TRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLP---------Y 117
TRIREY DFY VP+ LI K+ +++QM + PVQA + + V+ D+P Y
Sbjct 61 TRIREYLDFYFVPLRLINKNLPTALMQMQDN-PVQATGLSSNKVVTTDIPWVPTNLSGTY 119
Query 118 CSLSDLG-LSCFFASGSMSVPSLKSWQANNAYANIFGYIR 156
SL+ L + F S S + L + A A + Y+R
Sbjct 120 GSLTALADVKNSFPSSSTGIEDLLGFDAITQSAKLLMYLR 159
> Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1
Length=602
Score = 119 bits (298), Expect = 8e-33, Method: Compositional matrix adjust.
Identities = 69/187 (37%), Positives = 107/187 (57%), Gaps = 12/187 (6%)
Query 1 MAHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRP 60
MA+ ++N P RSGFD F+AK GELLPVYW + +PG ++++ FTRT P
Sbjct 1 MANLFSYGSVKNKPARSGFDWSEHFSFTAKAGELLPVYWKMLLPGTKVNLNLSSFTRTMP 60
Query 61 VQTAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSL 120
V TAAYTR++EY+D+Y VP+ LI KS +++QM + PVQA I+ +V+ DLP+ +
Sbjct 61 VNTAAYTRVKEYYDWYFVPLRLINKSIGQALVQM-QDQPVQATSIVANKSVTLDLPWTNA 119
Query 121 SDLGLSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNNMPAL 180
+ + +A+ ++ N Y N+ G+ + + KL+ L YGN + P+
Sbjct 120 ATMFTLLNYANVILT----------NKY-NLDGFSKAATSAKLLRYLRYGNCYYTSDPSK 168
Query 181 NIGNSNY 187
N N+
Sbjct 169 VGKNKNF 175
> Alpavirinae_Human_gut_33_005_Microviridae_AG0184_putative.VP1
Length=618
Score = 119 bits (297), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 63/179 (35%), Positives = 101/179 (56%), Gaps = 13/179 (7%)
Query 1 MAHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRP 60
MA ++N P RSGFD+G KN F+AK GELLPVYW +PG + I ++FTRT+P
Sbjct 1 MAGLFSYGDIKNKPRRSGFDLGNKNAFTAKVGELLPVYWKFCMPGDKFHISQEWFTRTQP 60
Query 61 VQTAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSL 120
V T+A+TRIREY++++ VP+ L++++ + +++ + E P A ++ + LP+ L
Sbjct 61 VDTSAFTRIREYYEWFFVPLHLMYRNSNEAIMSL-ENQPNYAASGTQSIVFNRKLPWVDL 119
Query 121 SDLGLSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNNMPA 179
L ++ + QA+ + N+FG+ R YKL + L G P+ A
Sbjct 120 QTLN------------DAITNVQASTYHNNMFGFARSSGFYKLFNSLGVGETDPSKTLA 166
> Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1
Length=584
Score = 117 bits (294), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 64/167 (38%), Positives = 96/167 (57%), Gaps = 18/167 (11%)
Query 7 LKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAY 66
L ++N P RSGFD+ +K FSAK GELLP+ W L +PG + + Q+FTRT+PV T+AY
Sbjct 6 LSSVKNRPRRSGFDLSSKVAFSAKVGELLPIKWTLTMPGDKFSLKEQHFTRTQPVNTSAY 65
Query 67 TRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSLSDLGLS 126
TR+REY+D++ P+ L+W++ + Q+ + A ++ + ++P
Sbjct 66 TRVREYYDWFWCPLHLLWRNAPEVIAQIQQNVQ-HASSFDGSVLLGSNMP---------- 114
Query 127 CFFASG-SMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNI 172
CF A S S+ +KS N FG+ R D+ YKLI L YGN+
Sbjct 115 CFSADQISQSLDMMKS------KLNYFGFNRADLAYKLIQYLRYGNV 155
> Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1
Length=618
Score = 115 bits (289), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 59/174 (34%), Positives = 102/174 (59%), Gaps = 13/174 (7%)
Query 1 MAHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRP 60
M+ ++N P RSGFD+ K F+AK GELLPVYW +PG ++I ++FTRT+P
Sbjct 1 MSSLFSYGDIKNSPRRSGFDLSNKCAFTAKVGELLPVYWKFCLPGDKFNISQEWFTRTQP 60
Query 61 VQTAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSL 120
V T+A+TRIREY++++ VP+ L++++ + +++ M E P A ++++ + +LP+ L
Sbjct 61 VDTSAFTRIREYYEWFFVPLHLLYRNSNEAIMSM-ENQPNYAASGSSSISFNRNLPWVDL 119
Query 121 SDLGLSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIP 174
S + + ++ + Q++ + N FG R + KL+ L YG P
Sbjct 120 STINV------------AIGNVQSSTSPKNFFGVSRSEGFKKLVSYLGYGETSP 161
Lambda K H a alpha
0.321 0.137 0.436 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 15519306