bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
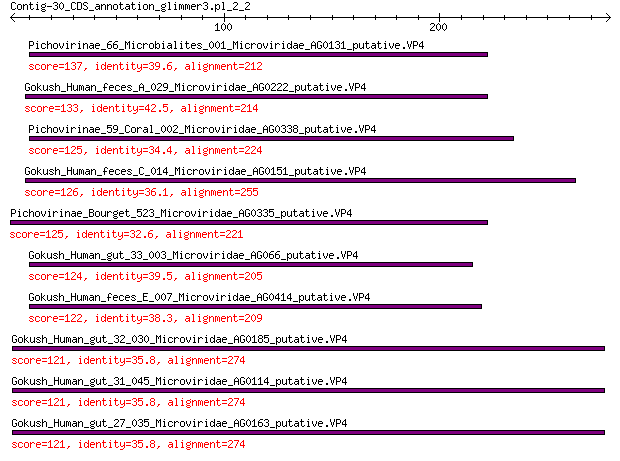
Query= Contig-30_CDS_annotation_glimmer3.pl_2_2
Length=278
Score E
Sequences producing significant alignments: (Bits) Value
Pichovirinae_66_Microbialites_001_Microviridae_AG0131_putative.VP4 137 5e-40
Gokush_Human_feces_A_029_Microviridae_AG0222_putative.VP4 133 5e-38
Pichovirinae_59_Coral_002_Microviridae_AG0338_putative.VP4 125 2e-35
Gokush_Human_feces_C_014_Microviridae_AG0151_putative.VP4 126 2e-35
Pichovirinae_Bourget_523_Microviridae_AG0335_putative.VP4 125 2e-35
Gokush_Human_gut_33_003_Microviridae_AG066_putative.VP4 124 6e-35
Gokush_Human_feces_E_007_Microviridae_AG0414_putative.VP4 122 4e-34
Gokush_Human_gut_32_030_Microviridae_AG0185_putative.VP4 121 1e-33
Gokush_Human_gut_31_045_Microviridae_AG0114_putative.VP4 121 1e-33
Gokush_Human_gut_27_035_Microviridae_AG0163_putative.VP4 121 1e-33
> Pichovirinae_66_Microbialites_001_Microviridae_AG0131_putative.VP4
Length=291
Score = 137 bits (346), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 84/215 (39%), Positives = 118/215 (55%), Gaps = 21/215 (10%)
Query 10 VPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSDLFQTNVAVV 69
VPCG C NC+ + WV+RL +K S FVTLTYD H+PI G F T +
Sbjct 35 VPCGRCPNCKLRRVNDWVFRLMQHSKVSDNSHFVTLTYDTRHVPITENG---FMT----L 87
Query 70 SKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGFPFTGKMAGDL--LAE 127
K V LFMKRLR KYE +++Y+ EYG N RPHYHMILFG D+ + +
Sbjct 88 DKDAVPLFMKRLR-KYETAQLKYYAVGEYGTNNKRPHYHMILFG-------VSDIENIHK 139
Query 128 CWQNGFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPGIGFGFMKAD 187
W G V +T IAY KY+ + P+ RD+ + F L S+ G+G ++ +
Sbjct 140 AWNYGSVWVGTVTGDSIAYTMKYIDKSRWRPDHWRDD-RIPEFPLMSK--GLGENYLTHE 196
Query 188 IIEFYRRH-PRDYVRAWAGHKMAMPRYYADKLYDD 221
++++++ R Y GHK+A+PRYY +K+Y D
Sbjct 197 VVKYHKSDVSRMYATKEGGHKIALPRYYRNKIYSD 231
> Gokush_Human_feces_A_029_Microviridae_AG0222_putative.VP4
Length=343
Score = 133 bits (335), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 91/250 (36%), Positives = 131/250 (52%), Gaps = 51/250 (20%)
Query 8 AKVPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSDLFQTNVA 67
++PCG C+ CR + W R E + + + FVT+TYDDEH+P + SD +T A
Sbjct 55 VQIPCGRCIGCRLEYSRQWANRCMLELQYHDSAYFVTVTYDDEHVP-QTYSSD-SETGEA 112
Query 68 VV-----SKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGF------PF 116
++ SKRD+QLFMKR+RK++ D ++RYF+ EYG+ RPHYH ILFG P+
Sbjct 113 LLPLMTLSKRDMQLFMKRVRKRFCDDRIRYFLAGEYGSTTFRPHYHCILFGLHLYDLVPY 172
Query 117 TGKMAGDL------LAECWQN------GFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDE 164
GD+ L+ CW + G+V P+T + AYV +Y +KS + D
Sbjct 173 AKNFRGDVLYNSQSLSACWCDKSARPMGYVVVAPVTYETCAYVARYTSKKSG----VNDL 228
Query 165 KKY------KPFMLCSRNPGIGFGFMKADIIEFYRRHPR----DYVRAW---AGHKMAMP 211
+ Y +PF L SR PGIG +++ HP D++ G K P
Sbjct 229 EAYDLLGLARPFTLMSRKPGIGR--------QYFDDHPDCMDYDFINVSTGDGGKKFHPP 280
Query 212 RYYADKLYDD 221
RYY +KLYD+
Sbjct 281 RYY-EKLYDE 289
> Pichovirinae_59_Coral_002_Microviridae_AG0338_putative.VP4
Length=274
Score = 125 bits (314), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 77/226 (34%), Positives = 117/226 (52%), Gaps = 15/226 (7%)
Query 10 VPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSDLFQTNVAVV 69
VPCG C+ C + + W +RL EAK + F+TLTY E+ P+ G F+T +
Sbjct 25 VPCGKCLACTKRRASQWSFRLNEEAKTSSSACFITLTY--ENAPVSENG---FRT----L 75
Query 70 SKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGFPFTGKMAGDLLAECW 129
+KRD QLF+KRLRKK K++Y+ EYG + RPHYH I+F P + + + W
Sbjct 76 NKRDFQLFLKRLRKKCPTNKLKYYACGEYGTRTFRPHYHAIIFNLPKSLIQDPQKIVDTW 135
Query 130 QNGFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKP-FMLCSRNPGIGFGFMKADI 188
+G + I YV YM + + E D+ P F L S+ +G G++ +
Sbjct 136 THGHIHLANNNQSTINYVVGYMTKGNF--ERFNDQDDRMPEFSLMSKK--MGMGYLTPQM 191
Query 189 IEFYRRHPRDYVRAWAGHKMAMPRYYADKLYD-DDMKAFLKEMREE 233
E+YR+ + GH ++MPRYY +K++ ++K K EE
Sbjct 192 KEYYRKREITCLVRENGHIISMPRYYKEKIFTKQELKKLYKRYIEE 237
> Gokush_Human_feces_C_014_Microviridae_AG0151_putative.VP4
Length=331
Score = 126 bits (316), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 92/274 (34%), Positives = 128/274 (47%), Gaps = 35/274 (13%)
Query 8 AKVPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSDLFQTNVA 67
A +PCG C CR+ W R++ EAK + +FVT+TYD+EH+P I
Sbjct 44 ALLPCGKCEYCRKQMADQWATRIELEAKRWDNVIFVTMTYDEEHIPYGEILKGYQSIQSQ 103
Query 68 VVSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGF-PFTGK------- 119
VSKRDVQLF+KRLRK Y+ ++YF+ EYG + RPHYH I FG P G
Sbjct 104 TVSKRDVQLFLKRLRKAYKK-PIKYFIAGEYGDRTKRPHYHGIFFGLKPEDGVWYKNQKG 162
Query 120 ---MAGDLLAECWQNGFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRN 176
+ L W GFV P AYV +Y+ +K++ E + + + R
Sbjct 163 NAYFKSEWLTNIWGKGFVDFSPAAPGSYAYVAQYVNKKAIGAE------QSAKYWMQGRE 216
Query 177 P-------GIGFGFMKADIIEFYRRHPRDYVRAWAGHKMAMPRYYADKLYDDDMKAFLKE 229
P GIG ++K + E D + G + PRY+ DKL D D E
Sbjct 217 PEFRIMSKGIGEEYLKEHMDEILE---TDNITCAGGRQKRPPRYF-DKLLDRDT----NE 268
Query 230 MREEFFR-HKMFNEWIDYCARENPILTDLMQLEQ 262
E +F+ H + R N +L+ L+ LEQ
Sbjct 269 DAESYFKAHSNELRAVRAKRRRNAVLS-LVNLEQ 301
> Pichovirinae_Bourget_523_Microviridae_AG0335_putative.VP4
Length=291
Score = 125 bits (314), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 72/224 (32%), Positives = 120/224 (54%), Gaps = 19/224 (8%)
Query 1 MYLASVEAKVPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSD 60
+Y + VPCG C C + W +RL+ AK S F+TLTYD ++PI G
Sbjct 23 IYSNDRQVPVPCGKCPACLSRRTSVWTFRLKTHAKNAISSYFITLTYDTRYVPISSRG-- 80
Query 61 LFQTNVAVVSKRDVQLFMKRLRKKY-EDYK-MRYFVTSEYGAKNGRPHYHMILFGFPFTG 118
+ KRDVQL+ KRLRK + +D++ ++Y++ EYG+K RPHYH+ILF
Sbjct 81 -----FLTLDKRDVQLYFKRLRKLHSKDHEPLKYYLAGEYGSKTFRPHYHIILFNANI-- 133
Query 119 KMAGDLLAECWQNGFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPG 178
+L+ + W G V LT AY KY+ + + P + +++ + F L S+
Sbjct 134 ----ELIHKAWDKGEVHIGELTEASAAYTAKYINKGKVIP-VHKNDDRLPEFSLMSKK-- 186
Query 179 IGFGFMKADIIEFYRRH-PRDYVRAWAGHKMAMPRYYADKLYDD 221
+G ++ II ++R R+++ G K+++PRY+ +K++ +
Sbjct 187 LGLNYLSEKIIRYHRSDIERNFITLEDGKKISLPRYFREKIWTE 230
> Gokush_Human_gut_33_003_Microviridae_AG066_putative.VP4
Length=318
Score = 124 bits (312), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 81/237 (34%), Positives = 118/237 (50%), Gaps = 36/237 (15%)
Query 10 VPCGWCVNCRQNKRQSWVYRLQAEAKEYPLS--LFVTLTYDDEHLP--IERIGSDLFQT- 64
+PCG C+ CR +R+ W R++ EA++YP F+TLTYDD+H+P I G + +
Sbjct 29 IPCGQCIGCRIRQREDWTTRIELEARDYPREQVWFITLTYDDDHVPGMIVNTGEIMRKVQ 88
Query 65 -----------NVAVVSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFG 113
+V ++ D+Q F+KRLRK Y K+RYF+ EYG + RPHYHMIL+G
Sbjct 89 YTWKPGEKRPESVQILLYTDIQKFLKRLRKAYRG-KLRYFIAGEYGEQTARPHYHMILYG 147
Query 114 FPFT-----------GKMAGDLLAECWQNG---FVQAHPLTIKEIA-YVCKYMYE-KSMC 157
+ T G LA W G QA P T + +A YV K MYE
Sbjct 148 WQPTDLEHLYKIQHNGYFTSKWLANLWGMGQIQIAQAVPETYRYVAGYVTKKMYEIDGQK 207
Query 158 PEILRDEKKYKPFMLCSRNPGIGFGFMKADIIEFYRRHPRDYVRAWAGHKMAMPRYY 214
+ + KPF S PG+G + + E ++ + Y++ G + +PRYY
Sbjct 208 ANTYYELGQTKPFACMSLKPGLGDHYYQEHKKEIWK---QGYIQCTNGKRAQIPRYY 261
> Gokush_Human_feces_E_007_Microviridae_AG0414_putative.VP4
Length=298
Score = 122 bits (306), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 80/241 (33%), Positives = 122/241 (51%), Gaps = 36/241 (15%)
Query 10 VPCGWCVNCRQNKRQSWVYRLQAEAKEYPLS--LFVTLTYDDEHLP--IERIGSDLFQT- 64
+PCG C+ CR +R+ W R++ EA++YP F+TLTY+D+++P I + G + +
Sbjct 9 IPCGQCIGCRIRQREDWTTRIELEARDYPKEQVWFITLTYNDDNVPGMIVKTGEIMRKVQ 68
Query 65 -----------NVAVVSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFG 113
+V ++ D+Q F+KRLRK Y+ K+RYFV EYG + RPHYHMIL+G
Sbjct 69 YTWKPGKKRPESVQILLYEDIQKFLKRLRKAYKG-KLRYFVAGEYGEQTARPHYHMILYG 127
Query 114 FPFT-----------GKMAGDLLAECWQNG---FVQAHPLTIKEIA-YVCKYMYE-KSMC 157
+ T G + L W G QA P T + +A YV K MYE
Sbjct 128 WEPTDLKNLYKIHHNGYYTSEWLENLWGMGQIQIAQAVPETYRYVAGYVTKKMYELDGKK 187
Query 158 PEILRDEKKYKPFMLCSRNPGIGFGFMKADIIEFYRRHPRDYVRAWAGHKMAMPRYYADK 217
+ + KPF S PG+G + + E +R + Y++ G + +PRYY +
Sbjct 188 ANAYYELGQTKPFACMSLKPGLGDHYYQEHKAEIWR---QGYIQCTNGKQAQIPRYYEKQ 244
Query 218 L 218
+
Sbjct 245 M 245
> Gokush_Human_gut_32_030_Microviridae_AG0185_putative.VP4
Length=310
Score = 121 bits (303), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 98/301 (33%), Positives = 142/301 (47%), Gaps = 43/301 (14%)
Query 2 YLASVEAKVPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSDL 61
Y S +PCG C+ CR + + W R E K + + F T TYDD+H+P R
Sbjct 20 YPRSALVPLPCGQCIGCRIDYSRQWANRCLLELKYHDSAWFCTFTYDDDHVP--RTYYPD 77
Query 62 FQTNVAV----VSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGF--- 114
+T A+ + KRD QL MKR+RKK+E+ K+R+F++ EYG++ RPHYH ILFG
Sbjct 78 PETGEAIPALTLQKRDFQLLMKRIRKKFENDKIRFFMSGEYGSQTFRPHYHAILFGLHLD 137
Query 115 ---PF-TGKMAGDL--------LAECWQNGFVQAHPLTIKEIAYVCKYMYEKSMCPE--I 160
P+ T K G+ L ECW G+V +T + AY +Y+ +K E
Sbjct 138 DLQPYKTVKEGGEYYTYYNSPSLQECWPYGYVVVGEVTWESCAYTARYVMKKLKGKEAKF 197
Query 161 LRDEKKYKPFMLCSRNPGIGFGFMKAD---IIEFYRRHPRDYVRAWAGHKMAMPRYYADK 217
D F L SR PGI + + + E Y + G K PRYY DK
Sbjct 198 YGDHNIQPEFSLMSRKPGIARQYFDENSHCVEEQY----INVSTPKGGKKFRPPRYY-DK 252
Query 218 LYD---DDMKAFLKEMREEFFRHKMFNEWIDYCARENPILTDLMQLEQREEYEKRMNERL 274
L+D + A LK +R + + M + + T L E R+ E++ + RL
Sbjct 253 LFDIECPEKSAELKSLRAKLAQQAMEAKLSN---------TSLDSYELRDVEEEKQSNRL 303
Query 275 R 275
+
Sbjct 304 K 304
> Gokush_Human_gut_31_045_Microviridae_AG0114_putative.VP4
Length=310
Score = 121 bits (303), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 98/301 (33%), Positives = 142/301 (47%), Gaps = 43/301 (14%)
Query 2 YLASVEAKVPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSDL 61
Y S +PCG C+ CR + + W R E K + + F T TYDD+H+P R
Sbjct 20 YPRSALVPLPCGQCIGCRIDYSRQWANRCLLELKYHDSAWFCTFTYDDDHVP--RTYYPD 77
Query 62 FQTNVAV----VSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGF--- 114
+T A+ + KRD QL MKR+RKK+E+ K+R+F++ EYG++ RPHYH ILFG
Sbjct 78 PETGEAIPALTLQKRDFQLLMKRIRKKFENDKIRFFMSGEYGSQTFRPHYHAILFGLHLD 137
Query 115 ---PF-TGKMAGDL--------LAECWQNGFVQAHPLTIKEIAYVCKYMYEKSMCPE--I 160
P+ T K G+ L ECW G+V +T + AY +Y+ +K E
Sbjct 138 DLQPYKTVKEGGEYYTYYNSPSLQECWPYGYVVVGEVTWESCAYTARYVMKKLKGKEAKF 197
Query 161 LRDEKKYKPFMLCSRNPGIGFGFMKAD---IIEFYRRHPRDYVRAWAGHKMAMPRYYADK 217
D F L SR PGI + + + E Y + G K PRYY DK
Sbjct 198 YGDHNIQPEFSLMSRKPGIARQYFDENSHCVEEQY----INVSTPKGGKKFRPPRYY-DK 252
Query 218 LYD---DDMKAFLKEMREEFFRHKMFNEWIDYCARENPILTDLMQLEQREEYEKRMNERL 274
L+D + A LK +R + + M + + T L E R+ E++ + RL
Sbjct 253 LFDIECPEKSAELKSLRAKLAQQAMEAKLSN---------TSLDSYELRDVEEEKQSNRL 303
Query 275 R 275
+
Sbjct 304 K 304
> Gokush_Human_gut_27_035_Microviridae_AG0163_putative.VP4
Length=310
Score = 121 bits (303), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 98/301 (33%), Positives = 142/301 (47%), Gaps = 43/301 (14%)
Query 2 YLASVEAKVPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSDL 61
Y S +PCG C+ CR + + W R E K + + F T TYDD+H+P R
Sbjct 20 YPRSALVPLPCGQCIGCRIDYSRQWANRCLLELKYHDSAWFCTFTYDDDHVP--RTYYPD 77
Query 62 FQTNVAV----VSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGF--- 114
+T A+ + KRD QL MKR+RKK+E+ K+R+F++ EYG++ RPHYH ILFG
Sbjct 78 PETGEAIPALTLQKRDFQLLMKRIRKKFENDKIRFFMSGEYGSQTFRPHYHAILFGLHLD 137
Query 115 ---PF-TGKMAGDL--------LAECWQNGFVQAHPLTIKEIAYVCKYMYEKSMCPE--I 160
P+ T K G+ L ECW G+V +T + AY +Y+ +K E
Sbjct 138 DLQPYKTVKEGGEYYTYYNSPSLQECWPYGYVVVGEVTWESCAYTARYVMKKLKGKEAKF 197
Query 161 LRDEKKYKPFMLCSRNPGIGFGFMKAD---IIEFYRRHPRDYVRAWAGHKMAMPRYYADK 217
D F L SR PGI + + + E Y + G K PRYY DK
Sbjct 198 YGDHNIQPEFSLMSRKPGIARQYFDENSHCVEEQY----INVSTPKGGKKFRPPRYY-DK 252
Query 218 LYD---DDMKAFLKEMREEFFRHKMFNEWIDYCARENPILTDLMQLEQREEYEKRMNERL 274
L+D + A LK +R + + M + + T L E R+ E++ + RL
Sbjct 253 LFDIECPEKSAELKSLRAKLAQQAMEAKLSN---------TSLDSYELRDVEEEKQSNRL 303
Query 275 R 275
+
Sbjct 304 K 304
Lambda K H a alpha
0.326 0.140 0.448 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 22981276