bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
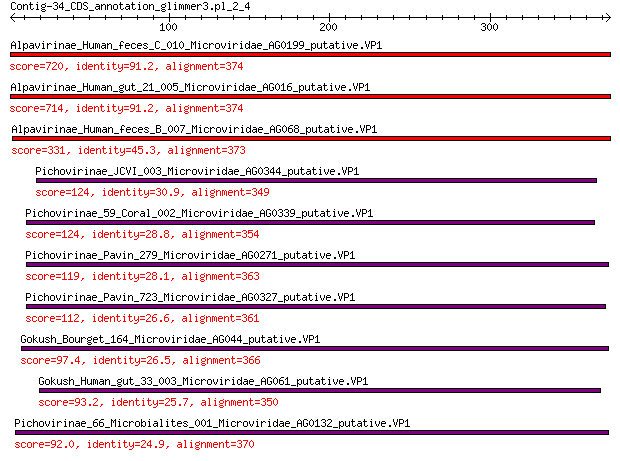
Query= Contig-34_CDS_annotation_glimmer3.pl_2_4
Length=374
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1 720 0.0
Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1 714 0.0
Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1 331 2e-107
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 124 2e-33
Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1 124 3e-33
Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1 119 2e-31
Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1 112 3e-29
Gokush_Bourget_164_Microviridae_AG044_putative.VP1 97.4 6e-24
Gokush_Human_gut_33_003_Microviridae_AG061_putative.VP1 93.2 2e-22
Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1 92.0 3e-22
> Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1
Length=731
Score = 720 bits (1859), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 341/374 (91%), Positives = 357/374 (95%), Gaps = 0/374 (0%)
Query 1 MKGGADNTTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQT 60
MKGGAD T Y L QCNWERDFLTTAVPNPQQGANAPLVGL +GDVVTR++DGTYS+QKQT
Sbjct 358 MKGGADTTLYELRQCNWERDFLTTAVPNPQQGANAPLVGLVMGDVVTRSEDGTYSVQKQT 417
Query 61 VLVDEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIE 120
VLVDEDGSKYG+SYKVSEDGE LVGVDYDPVSEKTPVTAI SYAELAAL T +GSGFTIE
Sbjct 418 VLVDEDGSKYGVSYKVSEDGERLVGVDYDPVSEKTPVTAIGSYAELAALATEQGSGFTIE 477
Query 121 TLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQT 180
TLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQT
Sbjct 478 TLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQT 537
Query 181 VDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLS 240
VDQQS SS+GQYAEALGSK+GIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQ+L
Sbjct 538 VDQQSGSSRGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQLLP 597
Query 241 KDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKY 300
KDF YNGLLDHYQPEFDRIGFQPITYKE+CP+N+ D+ N+ N+TFGYQRPWYEYVAKY
Sbjct 598 KDFTYNGLLDHYQPEFDRIGFQPITYKEICPMNIVAGDSANQLNKTFGYQRPWYEYVAKY 657
Query 301 DSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNAT 360
DSAHGLFRTNMKNF+MSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNAT
Sbjct 658 DSAHGLFRTNMKNFIMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNAT 717
Query 361 ARLPISRVAIPRLD 374
ARLPISRVAIPRLD
Sbjct 718 ARLPISRVAIPRLD 731
> Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1
Length=742
Score = 714 bits (1843), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 341/374 (91%), Positives = 356/374 (95%), Gaps = 1/374 (0%)
Query 1 MKGGADNTTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQT 60
MKGGAD T Y LHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTR++DGT S+QKQT
Sbjct 370 MKGGADETMYQLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRSEDGTLSVQKQT 429
Query 61 VLVDEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIE 120
VLVDEDGSKYGISYKVSEDGE LVGVDYDPVSEKTPVTAINSYAELAAL T +GSGFTIE
Sbjct 430 VLVDEDGSKYGISYKVSEDGERLVGVDYDPVSEKTPVTAINSYAELAALATEQGSGFTIE 489
Query 121 TLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQT 180
TLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQT
Sbjct 490 TLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQT 549
Query 181 VDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLS 240
VDQQS SSQGQYAEALGSK+GIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIY+Q+L
Sbjct 550 VDQQSASSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYSQLLP 609
Query 241 KDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKY 300
KDF YNGLLDHYQPEFDRIGFQPITYKEVCPLN +++ +N N+TFGYQRPWYEYVAKY
Sbjct 610 KDFTYNGLLDHYQPEFDRIGFQPITYKEVCPLNFDISN-MNDMNKTFGYQRPWYEYVAKY 668
Query 301 DSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNAT 360
D+AHGLFRT++K FVM R FSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNAT
Sbjct 669 DNAHGLFRTDLKKFVMHRTFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNAT 728
Query 361 ARLPISRVAIPRLD 374
ARLPISRVAIPRLD
Sbjct 729 ARLPISRVAIPRLD 742
> Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1
Length=780
Score = 331 bits (848), Expect = 2e-107, Method: Compositional matrix adjust.
Identities = 169/374 (45%), Positives = 245/374 (66%), Gaps = 16/374 (4%)
Query 2 KGGADN-TTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQT 60
+GG D+ T L NW+ D TTA+ PQQG APLVGLT + V+ D G T
Sbjct 422 EGGEDSLTPLDLLYANWQSDAYTTALTAPQQGV-APLVGLTTYESVSLNDAGHTVTTVNT 480
Query 61 VLVDEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIE 120
+VDEDG+ Y + ++ +GE+L GV+Y P+ AIN + +L + SG +I
Sbjct 481 AIVDEDGNAYKVDFE--SNGEALKGVNYTPLKAGE---AIN----MQSLVSPVTSGISIN 531
Query 121 TLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQT 180
R VNAYQ++LELN +GFSYK+I++GR+D+++R+D L MPE++GGI+R++ + + QT
Sbjct 532 DFRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQT 591
Query 181 VDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLS 240
V+ T+ G Y +LGS++G+A +G++ +I VFCDEES ++G++ V P+P+Y +L
Sbjct 592 VE---TTDTGSYVGSLGSQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILP 648
Query 241 KDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKY 300
K Y LD + PEFD IG+QPI KE+ + ++ N FGYQRPWYEYV K
Sbjct 649 KWLTYRERLDSFNPEFDHIGYQPIYLKELAAIQ--AFESGKDLNTVFGYQRPWYEYVQKV 706
Query 301 DSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNAT 360
D AHGLF ++++NF+M R F P+LG+ F ++ P +VN VFSVTE +DKI G + F+ T
Sbjct 707 DRAHGLFLSSLRNFIMFRSFENAPELGKSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCT 766
Query 361 ARLPISRVAIPRLD 374
A+LPISRV +PRL+
Sbjct 767 AQLPISRVVVPRLE 780
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 124 bits (311), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 108/357 (30%), Positives = 171/357 (48%), Gaps = 46/357 (13%)
Query 17 WERDFLTTAVPNPQQGANAPLVGLTVGDVVTRAD---DGTYSIQKQTVLVDEDGSKYGIS 73
+E D+ T+A+P Q+G NA V + +GD+ +D DG S+ K E+ + +S
Sbjct 181 FEHDYFTSALPFAQKG-NA--VDIPLGDITLNSDWYLDG--SVPKL-----ENSTGGYVS 230
Query 74 YKVSED---GESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVNAYQK 130
V+ D GE VG D DP + Y +L T TI LR Q+
Sbjct 231 GNVTSDNATGEINVGSDNDPFA----------YDPDGSLEVTST---TITDLRRAFKLQE 277
Query 131 FLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQG 190
+LEL R G Y + ++ +D+ L PE+I G + + + + T + + QG
Sbjct 278 WLELLARGGSRYIEQIKTFFDVKSSDQRLQRPEYITGTKQPVIISEILNTTGETAGLPQG 337
Query 191 QYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLD 250
+ SG GV T N + +C+E +IIG+L+VTP Y Q + K +L D
Sbjct 338 -------NMSG-HGVAVGTGNVGKYYCEEHGFIIGILSVTPNTAYQQGIPKHYLRTDKYD 389
Query 251 HYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTN 310
Y P+F IG QPI E+ + ++TFGY + EY G FR++
Sbjct 390 FYWPQFAHIGEQPIVNNEIYAYD-------PTGDETFGYTPRYAEYKYMPSRVAGEFRSS 442
Query 311 MKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYV--KFNATARLPI 365
+ + +R+FS LP L Q F+ VD D +++F+VT+ D ++ +V K A ++P+
Sbjct 443 LDEWHAARIFSSLPTLSQDFIEVDADDFDRIFAVTDGDDNLYMHVLNKVKARRQMPV 499
> Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1
Length=522
Score = 124 bits (311), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 102/366 (28%), Positives = 168/366 (46%), Gaps = 39/366 (11%)
Query 11 SLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLVDE----- 65
++ + W+ D+ T+A+P Q+G A + T + + T +I + +
Sbjct 177 TMRKRAWQHDYFTSALPWTQRGPEATIPLGTSAPITWSNNTATATIVRNNTSGNPIAGYT 236
Query 66 -DGSKY---GISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIET 121
DG+ G S ++ D + V +D+D + + +A+LA T + +I
Sbjct 237 FDGASALYTGGSGQLIADLPTPVSLDFD--------NSDHLFADLAGATAS-----SIND 283
Query 122 LRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQT- 180
LR Q++LE N R G Y +I+ + + L PEF+GG + +++ V QT
Sbjct 284 LRRAFRLQEWLERNARGGARYIEIIMAHFGVRSSDARLQRPEFLGGSATPITISEVLQTS 343
Query 181 VDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLS 240
+ + QG A GV +SN + C+E YIIG+++V P Y Q +
Sbjct 344 ANNTEPTPQGNMAGH--------GVSVGSSNYVSYKCEEHGYIIGIMSVMPKTAYQQGIP 395
Query 241 KDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKY 300
K F D+Y P F IG QPI +E+ N DT +TFGY + EY
Sbjct 396 KHFKKLDKFDYYWPSFANIGEQPILNEELYHQN-NATDT-----ETFGYTPRYAEYKYIP 449
Query 301 DSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYV--KFN 358
+ HG FR ++ + M R+F P L Q F+ + D V +VF+VT + ++ Y+ +
Sbjct 450 STVHGEFRDSLDFWHMGRIFGSKPTLNQDFIECNADDVERVFAVTAGQEHLYVYLHNEVK 509
Query 359 ATARLP 364
AT +P
Sbjct 510 ATRLMP 515
> Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1
Length=503
Score = 119 bits (297), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 102/369 (28%), Positives = 160/369 (43%), Gaps = 44/369 (12%)
Query 11 SLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQK-----QTVLVDE 65
+L + +E D+ T +P Q+GA V L +GDV +A+ +I K T + +
Sbjct 173 TLRKRAYEHDYFTACLPFAQKGA---AVDLPLGDVTLKANWNPSAIPKFVNAAGTSINNL 229
Query 66 DGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLRYV 125
D + G ++ G ++P+ A N L +TT I LR
Sbjct 230 DDVQGGTPNRIRTSGG------------QSPI-AYNPNGSLEVGSTT------INDLRRA 270
Query 126 NAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQS 185
Q+FLE N R G Y + ++ + + L PE+I G + ++ + TV
Sbjct 271 MRLQEFLEKNARGGTRYIENIKMHFGVQSSDSRLQRPEYITGTKAPVVVQEITSTVKGIE 330
Query 186 TSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLY 245
+A +SG G Y +E YIIG++++ P P Y Q + K +L
Sbjct 331 EPQGNPTGKATSLQSGHYGNYS---------VEEHGYIIGIMSIMPKPAYQQGIPKSYLK 381
Query 246 NGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHG 305
+LD Y P F IG Q + +E+ G T N FGY + EY + G
Sbjct 382 TDVLDFYWPSFANIGEQEVLQEEI----YGYTPT---PNAVFGYIPRYAEYKYLQNRVCG 434
Query 306 LFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTE-YTDKIFGYVKFNATARLP 364
LFRT + + + R+F+ +P L Q F+ V P ++F+ E Y D I+ +V TAR P
Sbjct 435 LFRTTLDFWHLGRIFANIPTLSQSFIEVSPANQTRIFANQEDYDDNIYIHVLNKMTARRP 494
Query 365 ISRVAIPRL 373
+ P L
Sbjct 495 MPVFGTPML 503
> Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1
Length=508
Score = 112 bits (281), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 96/363 (26%), Positives = 158/363 (44%), Gaps = 40/363 (11%)
Query 11 SLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLVDEDGSKY 70
+L Q WE D+ T ++P Q+GA V + +G + + V V+ + ++
Sbjct 181 TLRQRAWEHDYFTASLPFAQKGA---AVDIPIGTI------------DKDVAVNFNSLEF 225
Query 71 GISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVNAYQK 130
G + + L+ + V A + +A +T + TI LR Q+
Sbjct 226 GATTVRYNSADGLIDI-----GGSNAVNADGTPTMIAKTSTIDIEPTTINDLRRAFKLQE 280
Query 131 FLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQG 190
+LE N R G Y + + + + L PE+I G+ + + V T Q QG
Sbjct 281 WLEKNARGGTRYIENILTHFGVRSSDKRLQRPEYITGVKSPVVVSEVLNTTGQDGGLPQG 340
Query 191 QYA-EALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLL 249
A + SG +G Y +C+E YIIG+++V P Y Q + + FL L
Sbjct 341 NMAGHGISVTSGKSGSY---------YCEEHGYIIGIMSVMPKTAYQQGIPRTFLKTDSL 391
Query 250 DHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRT 309
D++ P F IG Q + +E+ A T N AN TFGY + EY G FRT
Sbjct 392 DYFWPTFANIGEQEVAKQEL------YAYTAN-ANDTFGYVPRYAEYKYMPSRVAGEFRT 444
Query 310 NMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTE-YTDKIFGYVKFNATARLPISRV 368
++ + + R+F+ P L F+ DP ++F+V + TD ++ +V A P+ +
Sbjct 445 SLNYWHLGRIFATEPSLNSDFIECDP--TKRIFAVEDPETDVLYCHVLNKIKAVRPMPKY 502
Query 369 AIP 371
P
Sbjct 503 GTP 505
> Gokush_Bourget_164_Microviridae_AG044_putative.VP1
Length=540
Score = 97.4 bits (241), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 97/372 (26%), Positives = 156/372 (42%), Gaps = 26/372 (7%)
Query 8 TTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLVDEDG 67
T Y++ + D+ T+A+P PQ+G A V + +G +G + T
Sbjct 185 TNYTILRRGKRHDYFTSALPWPQKGGTA--VTIPIGTSAPIKSNGGANFYAWTGSSGTSQ 242
Query 68 SKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVNA 127
K G S +++++G++ G D + T YA+L+ T TI LR
Sbjct 243 LKVGTSGQLTQNGQANPGSIVDQTWGPSGTTQ-GLYADLSTATAA-----TINQLRQSFQ 296
Query 128 YQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTS 187
QK LE + R G Y +I++ + + L PE++GG S +S+ + Q TS
Sbjct 297 IQKLLERDARGGTRYTEILRSHFGVTSPDARLQRPEYLGGGSTLVSISPIAQ------TS 350
Query 188 SQGQYAEALGSKSGIA-GVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYN 246
GQ +A + A G Y + ++ E YIIG+++V Y Q L + + +
Sbjct 351 GTGQTGQATPQGNLAAMGTYLAQNHGFTHSFVEHGYIIGVVSVRADLTYQQGLRRHWSRS 410
Query 247 GLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGL 306
D+Y P F +G Q I KE+ N N FGYQ W EY GL
Sbjct 411 TRYDYYFPAFAMLGEQAILNKEI------YVTGSNSDNNVFGYQERWAEYRYNPSEITGL 464
Query 307 FRTNMKNFV----MSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGY-VKFNATA 361
FR+ + ++ F+ LP L F+ +P + + + F FN A
Sbjct 465 FRSTAAGTIDPWHYAQRFTSLPTLNSTFIQDNPPLARNLAVGSAANGQQFLLDAFFNINA 524
Query 362 RLPISRVAIPRL 373
P+ ++P L
Sbjct 525 ARPLPMYSVPGL 536
> Gokush_Human_gut_33_003_Microviridae_AG061_putative.VP1
Length=545
Score = 93.2 bits (230), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 90/370 (24%), Positives = 163/370 (44%), Gaps = 45/370 (12%)
Query 19 RDFLTTAVPNPQQG--ANAPLVG---LTVGDVVTRADDGTYSIQKQTVLVDEDGSK---- 69
D+ T+++P Q+G P+ G + VG+ D ++ +++D G+
Sbjct 195 HDYFTSSLPFAQRGPEVTVPMTGNAPIRVGNAKGGYQDFRGPVE---MVIDAHGADIPGS 251
Query 70 --YGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVNA 127
YG S + +S++ + V+ + YA+++ +T TI LR A
Sbjct 252 LIYGNSTGAPGEKKSMMFSGKEKVTNEMGAGGW-MYADISEVTAA-----TINDLRKAVA 305
Query 128 YQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTS 187
Q++ E R G Y++ +Q WD+ I + +PE++GG ++M + QT QQ+
Sbjct 306 VQQYYEALARGGSRYREQVQALWDVVISDKTMQIPEYLGGGRYHVNMNQIVQTSGQQTND 365
Query 188 SQGQYAEALGSKSGIAGVYGSTSNNIEVFC---DEESYIIGLLTVTPVPIYTQMLSKDFL 244
+ G G T N F +E ++IG+ V Y Q L + +
Sbjct 366 D---------TPIGETGAMSVTPINESSFTKSFEEHGFVIGVCCVRHNHSYQQGLERFWS 416
Query 245 YNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAH 304
LD+Y P+F +G QPI KE+ + G A +TFGYQ W +Y K +
Sbjct 417 RTDRLDYYVPQFANLGEQPIKKKEI--MLTGTASD----EETFGYQEAWADYRMKPNRVS 470
Query 305 GLFRTNMKN----FVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKF--N 358
GL R+N + + + +S +P L Q+++ + + + + + + FG ++
Sbjct 471 GLMRSNAEGTLDFWHYADNYSTVPTLSQEWIAEGKEEIARTL-IVQNEPQFFGAIRIANK 529
Query 359 ATARLPISRV 368
T R+P+ V
Sbjct 530 TTRRMPLYSV 539
> Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1
Length=518
Score = 92.0 bits (227), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 92/376 (24%), Positives = 159/376 (42%), Gaps = 29/376 (8%)
Query 4 GADNTTYSLHQ-C---NWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQ 59
G +N+ +L+Q C W D+ T+ +P Q+G + L + + + + D T +
Sbjct 166 GDNNSQLALNQQCYRRAWMHDYFTSCLPFAQKGDSVQLPISSAENQIVQLDP-TATAPGT 224
Query 60 TVLVDEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTT-TEGSGFT 118
VL D S + + V D + P+ + +L +
Sbjct 225 FVLADGQASATPGNVHIINGPPPYVQTVVD--DDDNPL----QFEPRGSLVVDVQADAVD 278
Query 119 IETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVE 178
I T+R Q++LE N R G Y + + + + L PE++GG + + V
Sbjct 279 INTVRRAFRLQEWLEKNARAGTRYIESILSHFGVRSSDARLQRPEYLGGTKGNMVISEVL 338
Query 179 QTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQM 238
T + + A+G+ +G G+ N I+ C+E +IIG++ V PV Y Q
Sbjct 339 ATAETTDANV------AVGTMAG-HGITAHGGNTIKYRCEEHGWIIGIINVQPVTAYQQG 391
Query 239 LSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVA 298
L ++F LD+ P F IG Q + KE+ N D + FGY + E
Sbjct 392 LPREFSRFDRLDYPWPVFANIGEQEVYNKELY-ANSPAPDEI------FGYIPRYAEMKF 444
Query 299 KYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYT-DKIFGYVKF 357
K G R + + + R+F+ P L ++F+ +PDT ++F+VT+ D I ++
Sbjct 445 KNSRVAGEMRDTLDFWHLGRIFNNTPALNEEFIECNPDT--RIFAVTDPAEDHIVAHIYN 502
Query 358 NATARLPISRVAIPRL 373
T + R IP +
Sbjct 503 EVTVNRKLPRYGIPTI 518
Lambda K H a alpha
0.317 0.135 0.394 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 32586064