bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-35_CDS_annotation_glimmer3.pl_2_6
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
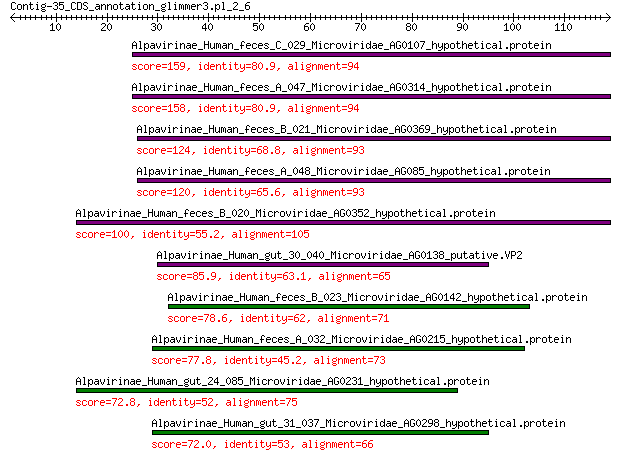
Alpavirinae_Human_feces_C_029_Microviridae_AG0107_hypothetical.... 159 1e-52
Alpavirinae_Human_feces_A_047_Microviridae_AG0314_hypothetical.... 158 2e-52
Alpavirinae_Human_feces_B_021_Microviridae_AG0369_hypothetical.... 124 4e-39
Alpavirinae_Human_feces_A_048_Microviridae_AG085_hypothetical.p... 120 9e-38
Alpavirinae_Human_feces_B_020_Microviridae_AG0352_hypothetical.... 100 8e-30
Alpavirinae_Human_gut_30_040_Microviridae_AG0138_putative.VP2 85.9 6e-24
Alpavirinae_Human_feces_B_023_Microviridae_AG0142_hypothetical.... 78.6 4e-21
Alpavirinae_Human_feces_A_032_Microviridae_AG0215_hypothetical.... 77.8 7e-21
Alpavirinae_Human_gut_24_085_Microviridae_AG0231_hypothetical.p... 72.8 6e-19
Alpavirinae_Human_gut_31_037_Microviridae_AG0298_hypothetical.p... 72.0 1e-18
> Alpavirinae_Human_feces_C_029_Microviridae_AG0107_hypothetical.protein
Length=105
Score = 159 bits (402), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 76/94 (81%), Positives = 86/94 (91%), Gaps = 0/94 (0%)
Query 25 RLKSVEIYEGESIETKCARILQNKEPITDTAPIIYTAKEDGVLPAYNIRTDRFDIAMDAY 84
R++S+EI EGE+IETK ARI QNKEPITD+APII+T K+DGVLPAYNIRTDRFDIA++A
Sbjct 12 RIQSIEIQEGETIETKVARITQNKEPITDSAPIIFTEKKDGVLPAYNIRTDRFDIALEAM 71
Query 85 DKITRSSAKKEIAPKPEDFGNVPNKTEGGSPSEN 118
DKI RS AKKE APKPEDFGNVPNKTEGG+PSEN
Sbjct 72 DKIGRSKAKKENAPKPEDFGNVPNKTEGGTPSEN 105
> Alpavirinae_Human_feces_A_047_Microviridae_AG0314_hypothetical.protein
Length=105
Score = 158 bits (400), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 76/94 (81%), Positives = 85/94 (90%), Gaps = 0/94 (0%)
Query 25 RLKSVEIYEGESIETKCARILQNKEPITDTAPIIYTAKEDGVLPAYNIRTDRFDIAMDAY 84
R++SVEI EGE+IETK ARI QNKEPITD+APII+T K+DGVLPAYNIRTDRFDIA++A
Sbjct 12 RIQSVEIQEGETIETKVARITQNKEPITDSAPIIFTEKKDGVLPAYNIRTDRFDIALEAM 71
Query 85 DKITRSSAKKEIAPKPEDFGNVPNKTEGGSPSEN 118
DKI RS AKKE PKPEDFGNVPNKTEGG+PSEN
Sbjct 72 DKIGRSKAKKENVPKPEDFGNVPNKTEGGTPSEN 105
> Alpavirinae_Human_feces_B_021_Microviridae_AG0369_hypothetical.protein
Length=93
Score = 124 bits (311), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 64/94 (68%), Positives = 75/94 (80%), Gaps = 2/94 (2%)
Query 26 LKSVEIYEGESIETKCARILQNKEPITDTAPIIYTAKEDGVLPAYNIRTDRFDIAMDAYD 85
+KSVE +EGE +E K RI+ N EPITD APII+T K+DGVLP YNIRTDR+DIA+DA +
Sbjct 1 MKSVECFEGEQLEEKVRRIVNNNEPITDGAPIIFTEKKDGVLPEYNIRTDRWDIALDAMN 60
Query 86 KITRS-SAKKEIAPKPEDFGNVPNKTEGGSPSEN 118
KI S A+KEI KPEDFGNVPNK + GSPSEN
Sbjct 61 KIDMSRKARKEIDVKPEDFGNVPNK-QNGSPSEN 93
> Alpavirinae_Human_feces_A_048_Microviridae_AG085_hypothetical.protein
Length=93
Score = 120 bits (302), Expect = 9e-38, Method: Compositional matrix adjust.
Identities = 61/94 (65%), Positives = 74/94 (79%), Gaps = 2/94 (2%)
Query 26 LKSVEIYEGESIETKCARILQNKEPITDTAPIIYTAKEDGVLPAYNIRTDRFDIAMDAYD 85
+KSVE +EGE +E K RI+ N EPITD APII+T K++GVLP YNIRTDR+DIA+DA D
Sbjct 1 MKSVECFEGEQLEEKVRRIVNNNEPITDGAPIIFTEKKNGVLPEYNIRTDRWDIALDAMD 60
Query 86 KITRS-SAKKEIAPKPEDFGNVPNKTEGGSPSEN 118
K+ + A+KE KPEDFGNVPNK + GSPSEN
Sbjct 61 KMEMARKARKETEVKPEDFGNVPNK-QNGSPSEN 93
> Alpavirinae_Human_feces_B_020_Microviridae_AG0352_hypothetical.protein
Length=102
Score = 100 bits (250), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 58/106 (55%), Positives = 73/106 (69%), Gaps = 5/106 (5%)
Query 14 MKYSFPTKNNDRLKSVEIYEGESIETKCARILQNKEPITDTAPIIYTAKEDGVLPAYNIR 73
MK + K R++SV YEGE+IE K RI+ N EPITD APIIYT ++DGVLP Y+IR
Sbjct 1 MKTAKLIKCVGRMESVTTYEGETIEAKVNRIVNNGEPITDGAPIIYTERKDGVLPEYDIR 60
Query 74 TDRFDIAMDAYDKITRSS-AKKEIAPKPEDFGNVPNKTEGGSPSEN 118
TDR+DIA+DA DK+ AK+E D +VP+K + GSPSEN
Sbjct 61 TDRWDIAIDAMDKVNMDRFAKRE---NKVDIKDVPDKKD-GSPSEN 102
> Alpavirinae_Human_gut_30_040_Microviridae_AG0138_putative.VP2
Length=111
Score = 85.9 bits (211), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 41/65 (63%), Positives = 50/65 (77%), Gaps = 0/65 (0%)
Query 30 EIYEGESIETKCARILQNKEPITDTAPIIYTAKEDGVLPAYNIRTDRFDIAMDAYDKITR 89
E +GESIETK RI +N EPITD APIIYT +EDGVLPAYNIRTDR++IA A + I +
Sbjct 16 EYQQGESIETKVKRITENNEPITDGAPIIYTNREDGVLPAYNIRTDRWEIAQAAMEAINQ 75
Query 90 SSAKK 94
++ K
Sbjct 76 TNLAK 80
> Alpavirinae_Human_feces_B_023_Microviridae_AG0142_hypothetical.protein
Length=110
Score = 78.6 bits (192), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 44/87 (51%), Positives = 56/87 (64%), Gaps = 16/87 (18%)
Query 32 YEGESIETKCARILQNKEPITDTAPIIYTAKEDGVLPAYNIRTDRFDIAMDAYD-----K 86
YEGE+IE K RI+ NKEPI D A IIYT K+DGVLP YNIRTD+++IA +A D +
Sbjct 19 YEGETIEHKVQRIVLNKEPIEDGAEIIYTEKKDGVLPQYNIRTDKWEIAQNAMDLAQQQR 78
Query 87 ITRSSA-----------KKEIAPKPED 102
I +S+ KKE PKP++
Sbjct 79 IAKSNGTYEAWHKENDKKKENEPKPDN 105
> Alpavirinae_Human_feces_A_032_Microviridae_AG0215_hypothetical.protein
Length=108
Score = 77.8 bits (190), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 51/73 (70%), Gaps = 0/73 (0%)
Query 29 VEIYEGESIETKCARILQNKEPITDTAPIIYTAKEDGVLPAYNIRTDRFDIAMDAYDKIT 88
VE YEG+SIE +C ++++ EPI DT+P+I+T KE GV+P Y++R D+++IA +A DK+
Sbjct 16 VESYEGQSIEDRCKKLVETGEPIKDTSPLIFTPKEKGVMPQYDVRADKWEIAQNAMDKVN 75
Query 89 RSSAKKEIAPKPE 101
+ K P E
Sbjct 76 KERIAKGQQPPAE 88
> Alpavirinae_Human_gut_24_085_Microviridae_AG0231_hypothetical.protein
Length=109
Score = 72.8 bits (177), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 39/75 (52%), Positives = 49/75 (65%), Gaps = 1/75 (1%)
Query 14 MKYSFPTKNNDRLKSVEIYEGESIETKCARILQNKEPITDTAPIIYTAKEDGVLPAYNIR 73
MK ++ K ++KS GE IE K AR+L +EPI D P+IYT K+ GV P YNIR
Sbjct 1 MKRTY-IKATKQIKSETPKRGEPIEIKVARLLSQEEPIKDQVPLIYTEKKTGVNPEYNIR 59
Query 74 TDRFDIAMDAYDKIT 88
TDRF IAM+A KI+
Sbjct 60 TDRFMIAMEAMGKIS 74
> Alpavirinae_Human_gut_31_037_Microviridae_AG0298_hypothetical.protein
Length=115
Score = 72.0 bits (175), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 35/67 (52%), Positives = 50/67 (75%), Gaps = 1/67 (1%)
Query 29 VEIYEGESIETKCARILQNKEPITDTAPIIYTAKEDGVLPAYNIRTDRFDIAMDAYDKIT 88
++ EGE IE K R++ K PI+D APIIYT ++DGVLPAY+IRTDR++IA A ++
Sbjct 18 ADLIEGERIEDKVRRLMDEKSPISDGAPIIYTERKDGVLPAYDIRTDRWEIAQKAMEENM 77
Query 89 RS-SAKK 94
++ SAK+
Sbjct 78 KAISAKR 84
Lambda K H a alpha
0.310 0.130 0.366 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 6697444