bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-37_CDS_annotation_glimmer3.pl_2_1
Length=112
Score E
Sequences producing significant alignments: (Bits) Value
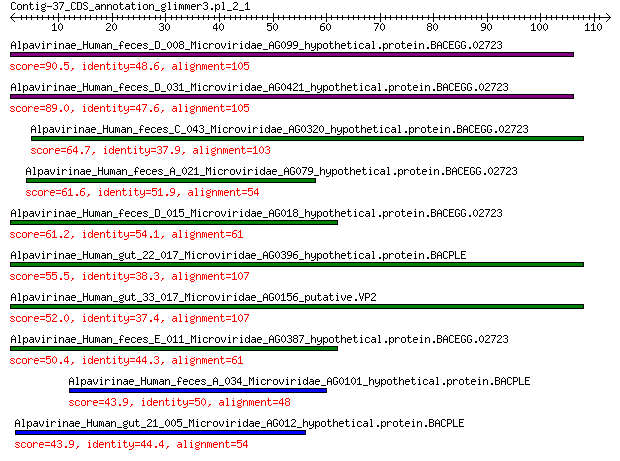
Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.p... 90.5 3e-24
Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.... 89.0 1e-23
Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.... 64.7 5e-15
Alpavirinae_Human_feces_A_021_Microviridae_AG079_hypothetical.p... 61.6 6e-14
Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.p... 61.2 9e-14
Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.p... 55.5 9e-12
Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2 52.0 1e-10
Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.... 50.4 5e-10
Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.... 43.9 6e-08
Alpavirinae_Human_gut_21_005_Microviridae_AG012_hypothetical.pr... 43.9 6e-08
> Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.protein.BACEGG.02723
Length=418
Score = 90.5 bits (223), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 51/114 (45%), Positives = 69/114 (61%), Gaps = 9/114 (8%)
Query 1 MNNQWNAEQAAINREWQTSERDAQNQWNLDQWNRENEYNSPAAQRARLEEAGYNPYMNGL 60
+NN++NA +A NR++QTSER+A QWNLDQWNREN YN P+AQRAR+E AG+NPY +
Sbjct 44 INNEFNASEALKNRDFQTSEREASQQWNLDQWNRENAYNDPSAQRARMEAAGFNPYNMNI 103
Query 61 DgntagtgvtsagvsgvgnPTAEMPNQVP--VAFNMDF-------SSIGNAINS 105
D +A T + TA +P + DF + IGNA++S
Sbjct 104 DAGSASTSGAQSSPGSGSQATASHTPSLPAYTGYAADFQNVASGIAQIGNAVSS 157
> Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.protein.BACEGG.02723
Length=418
Score = 89.0 bits (219), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 50/114 (44%), Positives = 68/114 (60%), Gaps = 9/114 (8%)
Query 1 MNNQWNAEQAAINREWQTSERDAQNQWNLDQWNRENEYNSPAAQRARLEEAGYNPYMNGL 60
+NN++NA +A NR++QTSER+A QWNLDQWNREN YN P+AQRAR+E AG+NPY +
Sbjct 44 INNEFNASEALKNRDFQTSEREASQQWNLDQWNRENAYNDPSAQRARMEAAGFNPYNMNI 103
Query 61 DgntagtgvtsagvsgvgnPTAEMPNQVP--VAFNMDF-------SSIGNAINS 105
D + T + + TA +P + DF + IGNA+ S
Sbjct 104 DPGSGSTSGAQSSPGSGSSATASHTPSLPAYTGYAADFQNVASGIAQIGNAVAS 157
> Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.protein.BACEGG.02723
Length=396
Score = 64.7 bits (156), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 39/105 (37%), Positives = 52/105 (50%), Gaps = 2/105 (2%)
Query 5 WNAEQAAINREWQTSERDAQNQWNLDQWNRENEYNSPAAQRARLEEAGYNP--YMNGLDg 62
+N +QA + R+W QN+WNL QWNREN YN+PAAQR+RLE AG N M G
Sbjct 48 FNEQQAQLGRDWSEEMMSQQNEWNLQQWNRENAYNTPAAQRSRLEAAGLNAALAMQGQGS 107
Query 63 ntagtgvtsagvsgvgnPTAEMPNQVPVAFNMDFSSIGNAINSYY 107
A A + P DFS + A++S++
Sbjct 108 IGMAGSGQPAAAPAGSPQAATGGSSAPQYSRPDFSLLSQAVDSFF 152
> Alpavirinae_Human_feces_A_021_Microviridae_AG079_hypothetical.protein.BACEGG.02723
Length=397
Score = 61.6 bits (148), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 28/54 (52%), Positives = 38/54 (70%), Gaps = 0/54 (0%)
Query 4 QWNAEQAAINREWQTSERDAQNQWNLDQWNRENEYNSPAAQRARLEEAGYNPYM 57
++ A+QA +NR +Q++E + +D WN+ NEYNS QRARLEEAG NPYM
Sbjct 7 KFQAQQAELNRNFQSAEAQKNRDFEVDMWNKTNEYNSATNQRARLEEAGLNPYM 60
> Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.protein.BACEGG.02723
Length=427
Score = 61.2 bits (147), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 33/61 (54%), Positives = 40/61 (66%), Gaps = 11/61 (18%)
Query 1 MNNQWNAEQAAINREWQTSERDAQNQWNLDQWNRENEYNSPAAQRARLEEAGYNPYMNGL 60
MNN++NA++A R +Q LD WN+EN YN+PAAQRARLEE GYN YMN
Sbjct 68 MNNEFNAKEAEKARAFQ-----------LDMWNKENAYNTPAAQRARLEEGGYNAYMNPA 116
Query 61 D 61
D
Sbjct 117 D 117
> Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.protein.BACPLE
Length=386
Score = 55.5 bits (132), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 41/109 (38%), Positives = 54/109 (50%), Gaps = 20/109 (18%)
Query 1 MNNQWNAEQAAINREWQTSERDAQNQWNLDQWNRENEYNSPAAQRARLEEAGYNPY--MN 58
MNNQ+N A R WQ + WN+EN YN+ +AQR RLEEAG NPY MN
Sbjct 41 MNNQFNERMAIQQRNWQE-----------NMWNKENAYNTASAQRQRLEEAGLNPYLMMN 89
Query 59 GLDgntagtgvtsagvsgvgnPTAEMPNQVPVAFNMDFSSIGNAINSYY 107
G A +G G+ + N V F D+S IG++I + +
Sbjct 90 G-------GSAGVAQSAGTGSAASSSGNAVMQPFQADYSGIGSSIGNIF 131
> Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2
Length=353
Score = 52.0 bits (123), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 40/109 (37%), Positives = 51/109 (47%), Gaps = 20/109 (18%)
Query 1 MNNQWNAEQAAINREWQTSERDAQNQWNLDQWNRENEYNSPAAQRARLEEAGYNPY--MN 58
MNNQ+N A R++Q + WN+EN YN+ +AQR RLEEAG NPY MN
Sbjct 41 MNNQFNERMAMQQRDFQE-----------NMWNKENTYNTASAQRQRLEEAGLNPYLMMN 89
Query 59 GLDgntagtgvtsagvsgvgnPTAEMPNQVPVAFNMDFSSIGNAINSYY 107
G + +G G + V F DFS I AI S +
Sbjct 90 G-------GSSGVSQSAGTGASASSSGTAVFQPFQADFSGIQQAIGSVF 131
> Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.protein.BACEGG.02723
Length=410
Score = 50.4 bits (119), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 39/61 (64%), Gaps = 12/61 (20%)
Query 1 MNNQWNAEQAAINREWQTSERDAQNQWNLDQWNRENEYNSPAAQRARLEEAGYNPYMNGL 60
MNN++NA +A R++Q+ + WN+ N++NSP R RL+EAGYNPY+ GL
Sbjct 82 MNNEFNAREAEKARQYQS-----------EMWNKTNDWNSPKNVRKRLQEAGYNPYL-GL 129
Query 61 D 61
D
Sbjct 130 D 130
> Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.protein.BACPLE
Length=365
Score = 43.9 bits (102), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 24/50 (48%), Positives = 30/50 (60%), Gaps = 2/50 (4%)
Query 12 INREWQTSERDAQNQWNLDQWNRENEYNSPAAQRARLEEAGYNPY--MNG 59
I EW +SE + ++ NE+NS +QRARLEEAG NPY MNG
Sbjct 39 IQNEWASSESQKSRDFAKSMFDASNEWNSAKSQRARLEEAGLNPYLMMNG 88
> Alpavirinae_Human_gut_21_005_Microviridae_AG012_hypothetical.protein.BACPLE
Length=383
Score = 43.9 bits (102), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 24/54 (44%), Positives = 31/54 (57%), Gaps = 0/54 (0%)
Query 2 NNQWNAEQAAINREWQTSERDAQNQWNLDQWNRENEYNSPAAQRARLEEAGYNP 55
N + +AE ++ ER AQ QW + + N YNSPAAQ RL+EAG NP
Sbjct 46 NRKQSAEAFERESKFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNP 99
Lambda K H a alpha
0.311 0.125 0.387 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 6160914