bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-3_CDS_annotation_glimmer3.pl_2_1
Length=233
Score E
Sequences producing significant alignments: (Bits) Value
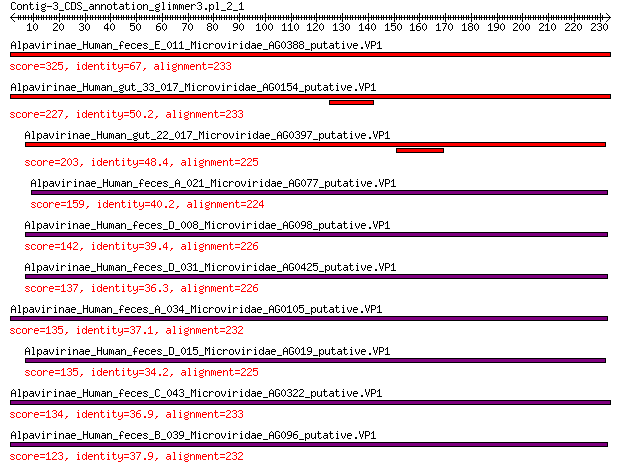
Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1 325 4e-109
Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1 227 2e-71
Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1 203 1e-62
Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1 159 1e-46
Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1 142 9e-41
Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1 137 7e-39
Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1 135 2e-38
Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1 135 3e-38
Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1 134 1e-37
Alpavirinae_Human_feces_B_039_Microviridae_AG096_putative.VP1 123 6e-34
> Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1
Length=622
Score = 325 bits (833), Expect = 4e-109, Method: Compositional matrix adjust.
Identities = 156/233 (67%), Positives = 188/233 (81%), Gaps = 1/233 (0%)
Query 1 MAHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRP 60
MAHFTGLKELQN PHK+G D+ +KN FTAKVGEL+PV+ DFAIP+C Y I+L YFTRTRP
Sbjct 1 MAHFTGLKELQNHPHKAGFDVGSKNLFTAKVGELIPVYWDFAIPDCDYDIDLAYFTRTRP 60
Query 61 VQTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSL 120
VQT+AYTR+REYFDF+AVPCDL+WKSFDSAVIQMG+ AP+QSK LL LTV D+P+C+L
Sbjct 61 VQTAAYTRVREYFDFYAVPCDLLWKSFDSAVIQMGQTAPVQSKTLLDPLTVGNDIPWCTL 120
Query 121 SDLSFALKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIG 180
DLS A+ F+SG+ + LG+ VSV GFGNIFGYNRGDV+ KLL L+YGN V+ S + +G
Sbjct 121 LDLSNAVYFSSGS-SPLGSTVSVPSGFGNIFGYNRGDVDSKLLFYLNYGNFVNPSLSNVG 179
Query 181 TNANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWEKC 233
+ +NRWWN +++ YSQKY N AVN+F L YQKIYQDFFRWSQWE
Sbjct 180 SPSNRWWNTSFSSSKVPGYSQKYLNNNAVNIFPLLAYQKIYQDFFRWSQWENA 232
> Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1
Length=614
Score = 227 bits (578), Expect = 2e-71, Method: Compositional matrix adjust.
Identities = 117/234 (50%), Positives = 154/234 (66%), Gaps = 9/234 (4%)
Query 1 MAHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRP 60
MA +TGL LQN PH+SG DI KN FTAKVGELLPV+ D ++P YR N+EYFTRT+P
Sbjct 1 MAFYTGLSNLQNHPHRSGFDIGRKNAFTAKVGELLPVYWDISMPGDKYRFNVEYFTRTQP 60
Query 61 VQTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSL 120
V TSAYTR+REYFDF+AVP L+WKS S + QM + IQ+ L NL++ LP ++
Sbjct 61 VATSAYTRLREYFDFYAVPLRLLWKSAPSVLTQMQDVNQIQALSLTQNLSLGTYLPSLTI 120
Query 121 SDLSFALKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIG 180
L +A+++ +GN + + N FG++R D++ KLLS L YGN+++ +
Sbjct 121 GTLGWAIRYLNGNTW----EPETASYLRNAFGFSRADLSFKLLSYLGYGNLIETPPSL-- 174
Query 181 TNANRWWNRRVA-TADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWEKC 233
NRWW+ + T D A Y+Q+Y N VN+F L TYQKIYQDFFRW QWEK
Sbjct 175 --GNRWWSTSLKNTDDGANYTQQYIQNTIVNIFPLLTYQKIYQDFFRWPQWEKS 226
Score = 21.2 bits (43), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 8/17 (47%), Positives = 13/17 (76%), Gaps = 0/17 (0%)
Query 125 FALKFASGNPASLGNKV 141
FAL ++ NP S+G+K+
Sbjct 325 FALDASTSNPVSVGSKL 341
> Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1
Length=607
Score = 203 bits (516), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 109/225 (48%), Positives = 141/225 (63%), Gaps = 10/225 (4%)
Query 7 LKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAY 66
+ LQN PH+SG DI KN FTAKVGELLPV+ D ++P Y+ N+EYFTRT+PV+TSAY
Sbjct 1 MSNLQNHPHRSGFDIGRKNAFTAKVGELLPVYWDISMPGDKYKFNVEYFTRTQPVETSAY 60
Query 67 TRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSLSDLSFA 126
TR+REYFDF+AVP L+WKS S + QM + +Q+ NL++ P +LS +
Sbjct 61 TRLREYFDFYAVPLRLLWKSAPSVLTQMQDINQLQALSFTQNLSLGSYFPSLTLSRFTAV 120
Query 127 LKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIGTNANRW 186
L +G GN S F N FG++R D+ KL S L YGNV +++NRW
Sbjct 121 LNRLNGGSNVPGN----SSTFLNEFGFSRADLAFKLFSYLGYGNVWSSEF----SSSNRW 172
Query 187 WNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWE 231
W+ + S Y+Q+Y + VNLF L YQKIYQDFFRWSQWE
Sbjct 173 WSTSLKGGGS--YTQQYVQDSYVNLFPLLAYQKIYQDFFRWSQWE 215
Score = 22.7 bits (47), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 9/18 (50%), Positives = 13/18 (72%), Gaps = 0/18 (0%)
Query 151 FGYNRGDVNHKLLSMLDY 168
FGY+ GDVN + +L+Y
Sbjct 542 FGYSEGDVNSQNKVVLNY 559
> Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1
Length=622
Score = 159 bits (401), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 90/224 (40%), Positives = 135/224 (60%), Gaps = 13/224 (6%)
Query 9 ELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAYTR 68
+L+N P +SG D++ + CFT+K GELLPV+ IP + I+ +FTRT+PV T+AYTR
Sbjct 8 DLKNHPRRSGFDLTKRICFTSKAGELLPVYWKPTIPGDKWNISTNWFTRTQPVDTAAYTR 67
Query 69 IREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSLSDLSFALK 128
++EY D+F VP +LI K +SA+ QM + P+ + + N + D+PY +L LS AL
Sbjct 68 VKEYVDWFFVPLNLIQKGIESAITQMVDN-PVSAMSAIENRAITTDMPYTTLLSLSRALY 126
Query 129 FASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIGTNANRWWN 188
+G + V+ G N+FG++R D++ KLL ML YGN ++ + + T +
Sbjct 127 MLNGK-----SYVNSHAGKLNMFGFSRADLSAKLLQMLKYGNFINPEHSGLDTPMFGY-- 179
Query 189 RRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWEK 232
+T A +S Y N AVN+ + YQKIY D+FR+ QWEK
Sbjct 180 ---STVKLAQFS--YLWNQAVNVLPIFCYQKIYSDYFRFQQWEK 218
> Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1
Length=618
Score = 142 bits (358), Expect = 9e-41, Method: Compositional matrix adjust.
Identities = 89/230 (39%), Positives = 131/230 (57%), Gaps = 16/230 (7%)
Query 7 LKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAY 66
+ ++N P +SG D+S + CFT+K GELLPV+ D P +++I + FTRT+P+ T+AY
Sbjct 6 MSAVKNHPRRSGFDLSNRVCFTSKAGELLPVFWDIVYPGDSFKIKTQLFTRTQPLNTAAY 65
Query 67 TRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSLSDLSFA 126
TRIREY DF+ VP LI K+ +A+ QM + P+Q+ L +N V D+P+ + S +
Sbjct 66 TRIREYLDFYFVPLRLINKNLPTALTQM-QDNPVQATGLSSNKVVTTDIPWVPVHSSSGS 124
Query 127 LKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGN----VVDKSANWIGTN 182
+G A + VS S N G++ + KLL L YGN VV IG +
Sbjct 125 YSSLTGF-ADVRGSVS-SSDIENFLGFDSITQSAKLLMYLRYGNFLSSVVPDEQKSIGLS 182
Query 183 ANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWEK 232
++ + +S S Y +V++ LATYQKIY DFFR++QWEK
Sbjct 183 SS------LDLRNSETVSTGYT---SVHILPLATYQKIYADFFRFTQWEK 223
> Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1
Length=614
Score = 137 bits (345), Expect = 7e-39, Method: Compositional matrix adjust.
Identities = 82/230 (36%), Positives = 129/230 (56%), Gaps = 15/230 (7%)
Query 7 LKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAY 66
+ ++N P +SG D+S + CFT+K GELLPV+ D P +++I + FTRT+P+ T+AY
Sbjct 1 MSAVKNHPRRSGFDLSNRVCFTSKAGELLPVFWDIVYPGDSFKIKTQLFTRTQPLNTAAY 60
Query 67 TRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSLSDLSFA 126
TRIREY DF+ VP LI K+ +A++QM + P+Q+ L +N V D+P+ +
Sbjct 61 TRIREYLDFYFVPLRLINKNLPTALMQM-QDNPVQATGLSSNKVVTTDIPWVPTN----- 114
Query 127 LKFASGNPASLGNK----VSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIGTN 182
L G+ +L + S S G ++ G++ + KLL L YGN + + N
Sbjct 115 LSGTYGSLTALADVKNSFPSSSTGIEDLLGFDAITQSAKLLMYLRYGNFLSSVVS--DKN 172
Query 183 ANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWEK 232
+ + + +S S Y ++++ LA YQK Y DFFR++QWEK
Sbjct 173 KSLGLSGSLDLRNSETASTGYT---SMHILPLAAYQKAYADFFRFTQWEK 219
> Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1
Length=618
Score = 135 bits (341), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 86/232 (37%), Positives = 126/232 (54%), Gaps = 39/232 (17%)
Query 1 MAHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRP 60
M+ +++N P +SG D+S K FTAKVGELLPV+ F +P + I+ E+FTRT+P
Sbjct 1 MSSLFSYGDIKNSPRRSGFDLSNKCAFTAKVGELLPVYWKFCLPGDKFNISQEWFTRTQP 60
Query 61 VQTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSL 120
V TSA+TRIREY+++F VP L++++ + A++ M E P + ++++ +LP+ L
Sbjct 61 VDTSAFTRIREYYEWFFVPLHLLYRNSNEAIMSM-ENQPNYAASGSSSISFNRNLPWVDL 119
Query 121 SDLSFALKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIG 180
S ++ A+ GN S S SP N FG +R + KL+S L YG
Sbjct 120 STINVAI----GNVQS-----STSP--KNFFGVSRSEGFKKLVSYLGYGETS-------- 160
Query 181 TNANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWEK 232
+KY NL + F L YQKIYQD++R SQWEK
Sbjct 161 -------------------PEKYVDNLRCSAFPLYAYQKIYQDYYRHSQWEK 193
> Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1
Length=584
Score = 135 bits (339), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 77/225 (34%), Positives = 128/225 (57%), Gaps = 20/225 (9%)
Query 7 LKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAY 66
L ++NRP +SG D+S+K F+AKVGELLP+ +P + + ++FTRT+PV TSAY
Sbjct 6 LSSVKNRPRRSGFDLSSKVAFSAKVGELLPIKWTLTMPGDKFSLKEQHFTRTQPVNTSAY 65
Query 67 TRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSLSDLSFA 126
TR+REY+D+F P L+W++ + Q+ + + ++ + ++P S +S +
Sbjct 66 TRVREYYDWFWCPLHLLWRNAPEVIAQIQQNVQ-HASSFDGSVLLGSNMPCFSADQISQS 124
Query 127 LKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIGTNANRW 186
L + +K+ N FG+NR D+ +KL+ L YGNV +GT+ +R
Sbjct 125 LDM-------MKSKL-------NYFGFNRADLAYKLIQYLRYGNV----RTGVGTSGSRN 166
Query 187 WNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWE 231
+ V DS+ Y+Q N A+++F + Y+K QD+FR +QW+
Sbjct 167 YGTSVDVKDSS-YNQNRAFNHALSVFPILAYKKFCQDYFRLTQWQ 210
> Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1
Length=602
Score = 134 bits (336), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 86/234 (37%), Positives = 131/234 (56%), Gaps = 18/234 (8%)
Query 1 MAHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRP 60
MA+ ++N+P +SG D S FTAK GELLPV+ +P +NL FTRT P
Sbjct 1 MANLFSYGSVKNKPARSGFDWSEHFSFTAKAGELLPVYWKMLLPGTKVNLNLSSFTRTMP 60
Query 61 VQTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSL 120
V T+AYTR++EY+D++ VP LI KS A++QM ++ P+Q+ ++ N +V DLP+ +
Sbjct 61 VNTAAYTRVKEYYDWYFVPLRLINKSIGQALVQMQDQ-PVQATSIVANKSVTLDLPWTNA 119
Query 121 SDLSFALKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKS-ANWI 179
+ + L +A+ L NK N+ G+++ + KLL L YGN S + +
Sbjct 120 ATMFTLLNYAN---VILTNKY-------NLDGFSKAATSAKLLRYLRYGNCYYTSDPSKV 169
Query 180 GTNANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWEKC 233
G N N + + D + + K N++ N+ LA YQKIY D+FR+ QWE
Sbjct 170 GKNKNFGLSSK---DDFNLLAAK---NVSFNVLPLAAYQKIYCDWFRFEQWENA 217
> Alpavirinae_Human_feces_B_039_Microviridae_AG096_putative.VP1
Length=579
Score = 123 bits (308), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 88/241 (37%), Positives = 125/241 (52%), Gaps = 30/241 (12%)
Query 1 MAHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRP 60
M+ F L + H+S D+S+K FTAKVGE+LP + AIP YRI+ ++FTRT P
Sbjct 1 MSDFNPLDRAKIPTHRSSFDLSSKKLFTAKVGEILPCYWQIAIPGTKYRISSDWFTRTVP 60
Query 61 VQTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSL 120
V T+AYTRI+EY+DF+AVP LI ++ A QM + + N + +P S+
Sbjct 61 VNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQMTDYM-TSAASSTANTSALTSVP--SV 117
Query 121 SDLSFALKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDK------ 174
+ F+ F + N N G ++G + KLL ML YG+++D
Sbjct 118 TQSLFSAFFQTANAGDQPNTRD-DAGLPIVYG------SCKLLDMLGYGSMIDSKNTGKA 170
Query 175 --SANWIGTNANRWWNRRVATADSA-VYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWE 231
+ ++G +A + AD+ VY VN L FLA YQKIY DFF +QWE
Sbjct 171 AITKKYLGVDA-------LGDADNPLVYQSSQTVNA---LPFLA-YQKIYYDFFSNNQWE 219
Query 232 K 232
K
Sbjct 220 K 220
Lambda K H a alpha
0.322 0.135 0.433 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 18284997