bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
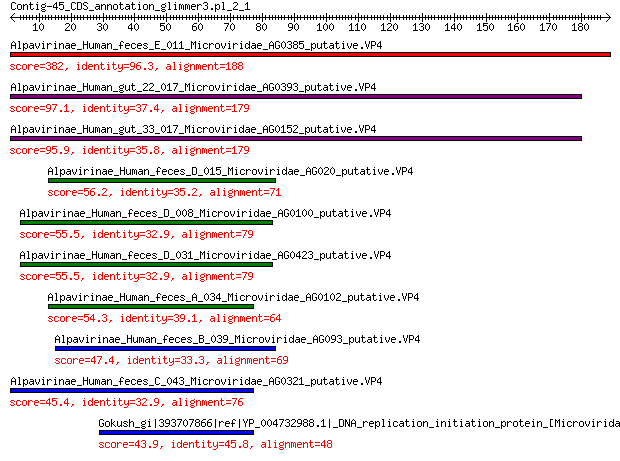
Query= Contig-45_CDS_annotation_glimmer3.pl_2_1
Length=188
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4 382 3e-132
Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4 97.1 4e-25
Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4 95.9 9e-25
Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4 56.2 2e-11
Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4 55.5 3e-11
Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4 55.5 4e-11
Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4 54.3 9e-11
Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4 47.4 2e-08
Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4 45.4 7e-08
Gokush_gi|393707866|ref|YP_004732988.1|_DNA_replication_initiat... 43.9 2e-07
> Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4
Length=611
Score = 382 bits (982), Expect = 3e-132, Method: Compositional matrix adjust.
Identities = 181/188 (96%), Positives = 183/188 (97%), Gaps = 0/188 (0%)
Query 1 MIPKEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHF 60
MIPKEELLTKYLFTECLRPQRIVNPYSHDVIFAPCG CKSCIMNKSNFATAYAMNMATHF
Sbjct 1 MIPKEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGHCKSCIMNKSNFATAYAMNMATHF 60
Query 61 KYCYFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRLLTPEYYHDRDLSL 120
KYCYFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPR LTPEYYHDRD SL
Sbjct 61 KYCYFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRFLTPEYYHDRDFSL 120
Query 121 DPAQNEVEQVFDIGFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILIKANG 180
D AQNEVEQVFD+GFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILIKANG
Sbjct 121 DSAQNEVEQVFDVGFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILIKANG 180
Query 181 RYDYGKNR 188
RYDYGK +
Sbjct 181 RYDYGKKK 188
> Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4
Length=562
Score = 97.1 bits (240), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 67/183 (37%), Positives = 86/183 (47%), Gaps = 31/183 (17%)
Query 1 MIPKEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHF 60
MI KE L L T C P+ +VN Y+H+ + CG C SCI+ +S+ T + F
Sbjct 1 MITKE--LQNKLVTRCQHPRTVVNKYTHEPVVVSCGHCPSCILRRSSVQTNLLTTYSAQF 58
Query 61 KYCYFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRLLTPEYYHDRDLSL 120
+Y YFVTLTY FLP L V VV T D D+S
Sbjct 59 RYVYFVTLTYAPSFLPTLEVSVVE---------------TCTDDI----------ADVSC 93
Query 121 DPAQNEVE----QVFDIGFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILI 176
P +E++ + GF+S+PR SVK K S R+F D +KF PMK EL IL
Sbjct 94 VPNIDELDASDPNTYLFGFRSVPRSSSVKLKSSTVERTFKDPDVKFSYPMKPKELLSILG 153
Query 177 KAN 179
K N
Sbjct 154 KIN 156
> Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4
Length=565
Score = 95.9 bits (237), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 64/179 (36%), Positives = 83/179 (46%), Gaps = 23/179 (13%)
Query 1 MIPKEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHF 60
MI KE L L T C P+ +VN Y+H+ + CG C SC++ +S T + F
Sbjct 1 MITKE--LQNKLVTRCQNPRTVVNKYTHEPVVVSCGACPSCVLRRSGIQTNLLTTYSAQF 58
Query 61 KYCYFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRLLTPEYYHDRDLSL 120
+Y YFVTLTY FLP L V V+ ++ + S D L
Sbjct 59 RYVYFVTLTYAPCFLPTLEVSVIETCT------DDIADVSSVPDIN------------DL 100
Query 121 DPAQNEVEQVFDIGFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILIKAN 179
DP N + GF S+PR SVK K S R+F D ++F PMK EL IL K N
Sbjct 101 DPCDN---NRYLFGFCSVPRSASVKLKNSTVERTFKDPEVRFSYPMKPKELLSILDKIN 156
> Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4
Length=563
Score = 56.2 bits (134), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 41/71 (58%), Gaps = 0/71 (0%)
Query 13 FTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYCYFVTLTYKD 72
F C PQ I N Y+ D + CG+C C++ K++ +T + + +YCYFVTLTY
Sbjct 9 FNRCEHPQVIQNKYTGDYVKVDCGQCPYCLIKKADRSTQKCDFVKYNHRYCYFVTLTYNT 68
Query 73 IFLPYLSVEVV 83
++P +S+ +
Sbjct 69 QYVPKMSLTQI 79
> Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4
Length=532
Score = 55.5 bits (132), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 26/79 (33%), Positives = 43/79 (54%), Gaps = 1/79 (1%)
Query 4 KEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYC 63
++E + K +F+ C + NPY+ ++I PCG C +C NKS + + ++
Sbjct 3 QQEFIQK-VFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLISRHV 61
Query 64 YFVTLTYKDIFLPYLSVEV 82
YFVTLTY ++PY E+
Sbjct 62 YFVTLTYAQRYIPYYEYEI 80
> Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4
Length=532
Score = 55.5 bits (132), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 26/79 (33%), Positives = 43/79 (54%), Gaps = 1/79 (1%)
Query 4 KEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYC 63
++E + K +F+ C + NPY+ ++I PCG C +C NKS + + ++
Sbjct 3 QQEFIQK-VFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLVSRHV 61
Query 64 YFVTLTYKDIFLPYLSVEV 82
YFVTLTY ++PY E+
Sbjct 62 YFVTLTYAQRYIPYYEYEI 80
> Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4
Length=539
Score = 54.3 bits (129), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 35/64 (55%), Gaps = 0/64 (0%)
Query 13 FTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYCYFVTLTYKD 72
+ CL P+ I N Y+ D I+ PCG C+ C+ NK+ A +YC F+TLTY
Sbjct 9 YGSCLHPRVIKNKYTGDPIYVPCGTCEFCVHNKAIKAELKCNVQLASSRYCEFITLTYST 68
Query 73 IFLP 76
+LP
Sbjct 69 EYLP 72
> Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4
Length=559
Score = 47.4 bits (111), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 35/69 (51%), Gaps = 0/69 (0%)
Query 15 ECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYCYFVTLTYKDIF 74
C I N Y+ I CG+C CI ++ A+ + FKY YFVTLTY +
Sbjct 13 HCQHRSFITNRYNGARITVDCGQCDYCIHKRAQKASMRVKTAGSAFKYSYFVTLTYDNEH 72
Query 75 LPYLSVEVV 83
+P ++ +V+
Sbjct 73 IPLMNCKVL 81
> Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4
Length=546
Score = 45.4 bits (106), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 25/79 (32%), Positives = 45/79 (57%), Gaps = 7/79 (9%)
Query 1 MIPKEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNM---A 57
M+P +++ K L CL + + N Y+ +++ CG+C++C+ S A+A ++ + A
Sbjct 1 MLPSDQI-HKELSKTCLHGKNVYNKYTGQMMYQSCGKCEACL---SRLASARSIKVGVQA 56
Query 58 THFKYCYFVTLTYKDIFLP 76
+ KY FVTLTY +P
Sbjct 57 SLSKYVMFVTLTYDTYHVP 75
> Gokush_gi|393707866|ref|YP_004732988.1|_DNA_replication_initiation_protein_[Microviridae_phi-CA82]
Length=353
Score = 43.9 bits (102), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 22/48 (46%), Positives = 27/48 (56%), Gaps = 0/48 (0%)
Query 29 DVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYCYFVTLTYKDIFLP 76
D I PCGRC C + KS A + +FKY YF+T+TY D LP
Sbjct 69 DGITVPCGRCHYCKLIKSYDRAKQAWCDSRYFKYQYFITITYDDDNLP 116
Lambda K H a alpha
0.325 0.140 0.427 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 13719484