bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-7_CDS_annotation_glimmer3.pl_2_9
Length=64
Score E
Sequences producing significant alignments: (Bits) Value
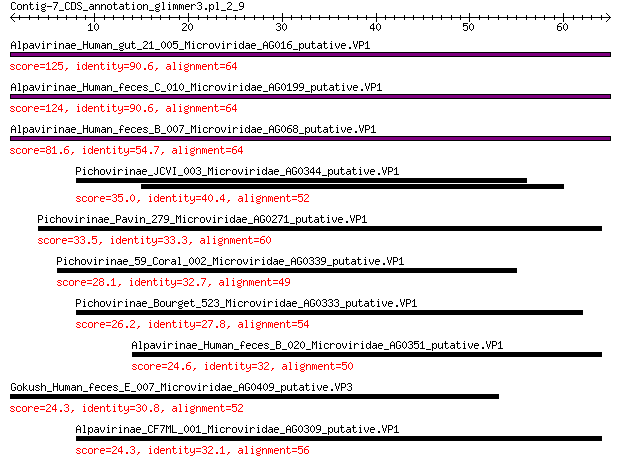
Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1 125 9e-37
Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1 124 3e-36
Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1 81.6 3e-21
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 35.0 2e-05
Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1 33.5 8e-05
Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1 28.1 0.004
Pichovirinae_Bourget_523_Microviridae_AG0333_putative.VP1 26.2 0.016
Alpavirinae_Human_feces_B_020_Microviridae_AG0351_putative.VP1 24.6 0.053
Gokush_Human_feces_E_007_Microviridae_AG0409_putative.VP3 24.3 0.080
Alpavirinae_CF7ML_001_Microviridae_AG0309_putative.VP1 24.3 0.084
> Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1
Length=742
Score = 125 bits (314), Expect = 9e-37, Method: Compositional matrix adjust.
Identities = 58/64 (91%), Positives = 62/64 (97%), Gaps = 0/64 (0%)
Query 1 MKNFVMHRSFTGLPQLGQQFLLVDPNAVNQVFSVTEYTDKIFGYVKFNATARLPISRVAI 60
+K FVMHR+F+GLPQLGQQFLLVDP+ VNQVFSVTEYTDKIFGYVKFNATARLPISRVAI
Sbjct 679 LKKFVMHRTFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAI 738
Query 61 PRLD 64
PRLD
Sbjct 739 PRLD 742
> Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1
Length=731
Score = 124 bits (311), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 58/64 (91%), Positives = 61/64 (95%), Gaps = 0/64 (0%)
Query 1 MKNFVMHRSFTGLPQLGQQFLLVDPNAVNQVFSVTEYTDKIFGYVKFNATARLPISRVAI 60
MKNF+M R F+GLPQLGQQFLLVDP+ VNQVFSVTEYTDKIFGYVKFNATARLPISRVAI
Sbjct 668 MKNFIMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAI 727
Query 61 PRLD 64
PRLD
Sbjct 728 PRLD 731
> Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1
Length=780
Score = 81.6 bits (200), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 35/64 (55%), Positives = 49/64 (77%), Gaps = 0/64 (0%)
Query 1 MKNFVMHRSFTGLPQLGQQFLLVDPNAVNQVFSVTEYTDKIFGYVKFNATARLPISRVAI 60
++NF+M RSF P+LG+ F ++ P +VN VFSVTE +DKI G + F+ TA+LPISRV +
Sbjct 717 LRNFIMFRSFENAPELGKSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVV 776
Query 61 PRLD 64
PRL+
Sbjct 777 PRLE 780
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 35.0 bits (79), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 31/50 (62%), Gaps = 2/50 (4%)
Query 8 RSFTGLPQLGQQFLLVDPNAVNQVFSVTEYTDKIFGYV--KFNATARLPI 55
R F+ LP L Q F+ VD + +++F+VT+ D ++ +V K A ++P+
Sbjct 450 RIFSSLPTLSQDFIEVDADDFDRIFAVTDGDDNLYMHVLNKVKARRQMPV 499
Score = 21.6 bits (44), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 13/45 (29%), Positives = 21/45 (47%), Gaps = 5/45 (11%)
Query 15 QLGQQFLLVDPNAVNQVFSVTEYTDKIFGYVKFNATARLPISRVA 59
+G+Q P N++++ D+ FGY A + SRVA
Sbjct 397 HIGEQ-----PIVNNEIYAYDPTGDETFGYTPRYAEYKYMPSRVA 436
> Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1
Length=503
Score = 33.5 bits (75), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 20/61 (33%), Positives = 30/61 (49%), Gaps = 1/61 (2%)
Query 4 FVMHRSFTGLPQLGQQFLLVDPNAVNQVFSVTE-YTDKIFGYVKFNATARLPISRVAIPR 62
+ + R F +P L Q F+ V P ++F+ E Y D I+ +V TAR P+ P
Sbjct 443 WHLGRIFANIPTLSQSFIEVSPANQTRIFANQEDYDDNIYIHVLNKMTARRPMPVFGTPM 502
Query 63 L 63
L
Sbjct 503 L 503
> Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1
Length=522
Score = 28.1 bits (61), Expect = 0.004, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 27/51 (53%), Gaps = 2/51 (4%)
Query 6 MHRSFTGLPQLGQQFLLVDPNAVNQVFSVTEYTDKIFGYV--KFNATARLP 54
M R F P L Q F+ + + V +VF+VT + ++ Y+ + AT +P
Sbjct 465 MGRIFGSKPTLNQDFIECNADDVERVFAVTAGQEHLYVYLHNEVKATRLMP 515
> Pichovirinae_Bourget_523_Microviridae_AG0333_putative.VP1
Length=518
Score = 26.2 bits (56), Expect = 0.016, Method: Composition-based stats.
Identities = 15/55 (27%), Positives = 27/55 (49%), Gaps = 3/55 (5%)
Query 8 RSFTGLPQLGQQFLLVDPNAVNQVFSVTEY-TDKIFGYVKFNATARLPISRVAIP 61
R F+ P L F+ DP+ ++F+V + D I+G++ N A + + P
Sbjct 464 RKFSAAPNLNGAFIECDPS--TRIFAVEDAEVDNIYGHIFNNIKAIRKMPKYGTP 516
> Alpavirinae_Human_feces_B_020_Microviridae_AG0351_putative.VP1
Length=651
Score = 24.6 bits (52), Expect = 0.053, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 24/51 (47%), Gaps = 1/51 (2%)
Query 14 PQLGQQFLLVDPNAVNQVFSVTEYTDKIFGY-VKFNATARLPISRVAIPRL 63
P++ +DP N +F+ T F +KF+ TAR +S IP L
Sbjct 601 PRIADLTTYIDPIKYNYIFADTSIDAMNFWVQIKFDITARRLMSAKQIPNL 651
> Gokush_Human_feces_E_007_Microviridae_AG0409_putative.VP3
Length=182
Score = 24.3 bits (51), Expect = 0.080, Method: Compositional matrix adjust.
Identities = 16/52 (31%), Positives = 25/52 (48%), Gaps = 4/52 (8%)
Query 1 MKNFVMHRSFTGLPQLGQQFLLVDPNAVNQVFSVTEYTDKIFGYVKFNATAR 52
+KN V SF P+ + LVD +++ +TE+ I Y ATA+
Sbjct 108 IKNIVRKASFD--PEFAKS--LVDSAKTDEIIDITEWPTNIHEYHAMIATAQ 155
> Alpavirinae_CF7ML_001_Microviridae_AG0309_putative.VP1
Length=692
Score = 24.3 bits (51), Expect = 0.084, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 27/60 (45%), Gaps = 7/60 (12%)
Query 8 RSFTGLPQLGQQFLLVDPNAVNQVFSV----TEYTDKIFGYVKFNATARLPISRVAIPRL 63
+ + G+P FL VDP N+V V TE TD +F+ +S +PR+
Sbjct 636 KEYKGVPT---NFLFVDPAVTNEVVEVNYDGTEKTDPFRISSRFSVQYISDMSVSGMPRV 692
Lambda K H a alpha
0.327 0.140 0.410 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3692164