bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-11_CDS_annotation_glimmer3.pl_2_1
Length=119
Score E
Sequences producing significant alignments: (Bits) Value
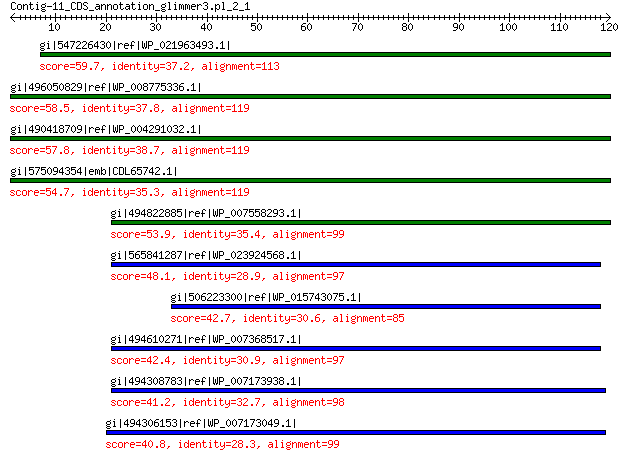
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 59.7 6e-08
gi|496050829|ref|WP_008775336.1| hypothetical protein 58.5 1e-07
gi|490418709|ref|WP_004291032.1| hypothetical protein 57.8 3e-07
gi|575094354|emb|CDL65742.1| unnamed protein product 54.7 3e-06
gi|494822885|ref|WP_007558293.1| hypothetical protein 53.9 5e-06
gi|565841287|ref|WP_023924568.1| hypothetical protein 48.1 4e-04
gi|506223300|ref|WP_015743075.1| hypothetical protein 42.7 0.013
gi|494610271|ref|WP_007368517.1| capsid protein 42.4 0.029
gi|494308783|ref|WP_007173938.1| hypothetical protein 41.2 0.075
gi|494306153|ref|WP_007173049.1| hypothetical protein 40.8 0.084
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 59.7 bits (143), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 42/123 (34%), Positives = 60/123 (49%), Gaps = 15/123 (12%)
Query 7 LNLQNNPGRNVSGALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPL-DGW-------- 57
L++ P S +GY RY K++ID +H F SWV+PL D +
Sbjct 456 FGLEDLPSDPSSINMGYVPRYADLKTSIDEIHGSFIDTLV--SWVSPLTDSYISAYRQAC 513
Query 58 -NVLTSSGAWSYQSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNG 116
+ S +Y KV P +++IF + DS ++ DQLL N F + AV+N D NG
Sbjct 514 KDAGFSDITMTYNFFKVNPHIVDNIFGVKADS---TINTDQLLINSYFDIKAVRNFDYNG 570
Query 117 LPY 119
LPY
Sbjct 571 LPY 573
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 58.5 bits (140), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 45/133 (34%), Positives = 68/133 (51%), Gaps = 19/133 (14%)
Query 1 MQSVPSLNLQN--NPGRNV-SGALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDGW 57
M+SVP ++L N NV S LGY RY +K+++D+ F+ +SWV D
Sbjct 453 MESVPLVSLMNPLQSSYNVGSSILGYAPRYISYKTDVDSSVGAFKT--TLKSWVMSYDNQ 510
Query 58 NVLT----------SSGAW-SYQSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQV 106
+V+ S G +Y + KV P ++ +F +A+ S+ DQ LC+ F V
Sbjct 511 SVINQLNYQDDPNNSPGTLVNYTNFKVNPNCVDPLFAV---AASNSIDTDQFLCSSFFDV 567
Query 107 YAVQNLDRNGLPY 119
V+NLD +GLPY
Sbjct 568 KVVRNLDTDGLPY 580
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 57.8 bits (138), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 46/142 (32%), Positives = 65/142 (46%), Gaps = 30/142 (21%)
Query 1 MQSVPSLNLQNNPGRNVSGA----LGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVA---- 52
MQS+P + L N P R+ + A LGY RY +K+++D GF+ SWV
Sbjct 444 MQSMPLVQLMN-PLRSFANASGLVLGYVPRYIDYKTSVDQSVGGFKR--TLNSWVISYGN 500
Query 53 --------------PLDGWNVLTSSGAWSYQSMKVRPQQLNSIFVPQV-DSANCSVAFDQ 97
P++ + S ++ KV P L+ IF Q D N DQ
Sbjct 501 ISVLKQVTLPNDAPPIEPSEPVPSVAPMNFTFFKVNPDCLDPIFAVQAGDDTNT----DQ 556
Query 98 LLCNVNFQVYAVQNLDRNGLPY 119
LC+ F + AV+NLD +GLPY
Sbjct 557 FLCSSFFDIKAVRNLDTDGLPY 578
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 54.7 bits (130), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 42/136 (31%), Positives = 65/136 (48%), Gaps = 23/136 (17%)
Query 1 MQSVPSLNLQNNPGRNVSGA----LGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDG 56
M+SVP + N + + + LGY RY WK+++D F + ++W P+ G
Sbjct 486 MESVPLVRAMNPVKESDTPSADTFLGYAPRYIDWKTSVDRSVGDF--ADSLRTWCLPV-G 542
Query 57 WNVLTSSGAWSYQS-------------MKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVN 103
LTS+ + ++ S KV P ++ +F DS +V D+ LC+
Sbjct 543 DKELTSANSLNFPSNPNVEPDSIAAGFFKVNPSIVDPLFAVVADS---TVKTDEFLCSSF 599
Query 104 FQVYAVQNLDRNGLPY 119
F V V+NLD NGLPY
Sbjct 600 FDVKVVRNLDVNGLPY 615
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 53.9 bits (128), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 35/111 (32%), Positives = 55/111 (50%), Gaps = 17/111 (15%)
Query 21 LGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDGWNVLTSSG----------AWSYQS 70
GY +Y+ WK+ +D FR + ++W+ P D +L + A S ++
Sbjct 508 FGYAPQYYNWKTTLDKSMGEFRR--SLKTWIIPFDDEALLAADSVDFPDNPNVEADSVKA 565
Query 71 --MKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY 119
KV P L+++F + AN + DQ LC+ F V V++LD NGLPY
Sbjct 566 GFFKVSPSVLDNLFAVK---ANSDLNTDQFLCSTLFDVNVVRSLDPNGLPY 613
>gi|565841287|ref|WP_023924568.1| hypothetical protein [Prevotella nigrescens]
gi|564729907|gb|ETD29851.1| hypothetical protein HMPREF1173_00033 [Prevotella nigrescens
CC14M]
Length=656
Score = 48.1 bits (113), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/97 (29%), Positives = 47/97 (48%), Gaps = 5/97 (5%)
Query 21 LGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDGWNVLTSSGAWSYQSMKVRPQQLNS 80
LGY+ RY ++K+ D + F +G + +W P + + G S + V P+ L
Sbjct 562 LGYSARYLEYKTARDIIFGEFMSGGSLSAWATPKNNYTF--EFGKLSLPDLLVDPKVLEP 619
Query 81 IFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGL 117
IF + N S++ DQ L N F V A++ + N +
Sbjct 620 IFAVKY---NGSMSTDQFLVNSYFDVKAIRPMQVNDM 653
>gi|506223300|ref|WP_015743075.1| hypothetical protein [Candidatus Methylomirabilis oxyfera]
gi|392373567|ref|YP_003205400.1| hypothetical protein DAMO_0481 [Candidatus Methylomirabilis oxyfera]
gi|258591260|emb|CBE67557.1| protein of unknown function [Candidatus Methylomirabilis oxyfera]
Length=234
Score = 42.7 bits (99), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 26/85 (31%), Positives = 43/85 (51%), Gaps = 1/85 (1%)
Query 33 NIDTVHAGFRAGAAYQSWVAPLDGWNVLTSSGAWSYQSMKVRPQQLNSIFVPQVDSANCS 92
+I+ V A R GAA+ + PL VL S A +++ K+R Q + V + +
Sbjct 86 SIENVIAAMRRGAAFDYLLKPLQDLTVLEVSVARAFEIRKLRAQAREAFQVGAIRELAVT 145
Query 93 VAFDQLLCNVNFQVYAVQNLDRNGL 117
A D++L +N +V+ L RNG+
Sbjct 146 -ASDRILNPLNIISLSVERLTRNGM 169
>gi|494610271|ref|WP_007368517.1| capsid protein [Prevotella multiformis]
gi|324988543|gb|EGC20506.1| putative capsid protein (F protein) [Prevotella multiformis DSM
16608]
Length=531
Score = 42.4 bits (98), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 30/104 (29%), Positives = 47/104 (45%), Gaps = 13/104 (13%)
Query 21 LGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDGWNVLTSSG-------AWSYQSMKV 73
LG+ +RY ++K++ D V F +G + W +P + +G WS V
Sbjct 431 LGWQVRYNEYKTSRDLVFGEFESGLSLSYWCSPRYDFGFDGKAGDKKLVNSPWSPAHFYV 490
Query 74 RPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGL 117
P LN+IF+ +V D L N F V AV+ + +GL
Sbjct 491 NPSILNTIFLV------SAVKADHFLVNSFFDVKAVRPMSVSGL 528
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 41.2 bits (95), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 32/99 (32%), Positives = 46/99 (46%), Gaps = 7/99 (7%)
Query 21 LGYNLRYWQWKSNIDTVHAGFRAGAAYQSW-VAPLDGWNVLTSSGAWSYQSMKVRPQQLN 79
LGY RY ++K+ +D H F A SW V+ W T+ K+ P LN
Sbjct 459 LGYQPRYSEYKTALDVNHGQFAQSDALSSWSVSRFRRW---TTFPQLEIADFKIDPGCLN 515
Query 80 SIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGLP 118
SIF VD N + A D + NF + V ++ +G+P
Sbjct 516 SIF--PVD-YNGTEANDCVYGGCNFNIVKVSDMSVDGMP 551
>gi|494306153|ref|WP_007173049.1| hypothetical protein [Prevotella bergensis]
gi|270333881|gb|EFA44667.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=519
Score = 40.8 bits (94), Expect = 0.084, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 45/100 (45%), Gaps = 7/100 (7%)
Query 20 ALGYNLRYWQWKSNIDTVHAGFRAGAAYQSW-VAPLDGWNVLTSSGAWSYQSMKVRPQQL 78
LGY RY ++K+ +D H F A SW V+ W T+ K+ P L
Sbjct 424 VLGYQPRYSEYKTALDINHGQFAQNDALSSWSVSRFRRW---TTFPQLEIADFKIDPGCL 480
Query 79 NSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGLP 118
NS+F + N + + D + NF + V ++ +G+P
Sbjct 481 NSVFPVEF---NGTESTDCVFGGCNFNIVKVSDMSVDGMP 517
Lambda K H a alpha
0.318 0.131 0.423 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 440495117073