bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-12_CDS_annotation_glimmer3.pl_2_3
Length=369
Score E
Sequences producing significant alignments: (Bits) Value
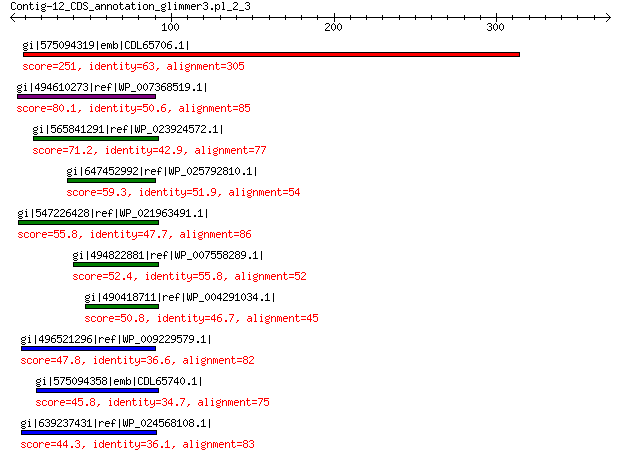
gi|575094319|emb|CDL65706.1| unnamed protein product 251 7e-76
gi|494610273|ref|WP_007368519.1| hypothetical protein 80.1 3e-13
gi|565841291|ref|WP_023924572.1| hypothetical protein 71.2 2e-10
gi|647452992|ref|WP_025792810.1| hypothetical protein 59.3 2e-06
gi|547226428|ref|WP_021963491.1| putative uncharacterized protein 55.8 2e-05
gi|494822881|ref|WP_007558289.1| hypothetical protein 52.4 2e-04
gi|490418711|ref|WP_004291034.1| hypothetical protein 50.8 8e-04
gi|496521296|ref|WP_009229579.1| hypothetical protein 47.8 0.009
gi|575094358|emb|CDL65740.1| unnamed protein product 45.8 0.029
gi|639237431|ref|WP_024568108.1| hypothetical protein 44.3 0.071
>gi|575094319|emb|CDL65706.1| unnamed protein product [uncultured bacterium]
Length=396
Score = 251 bits (641), Expect = 7e-76, Method: Compositional matrix adjust.
Identities = 192/306 (63%), Positives = 237/306 (77%), Gaps = 1/306 (0%)
Query 9 SLLSPG-ISAIGSIVGGLFGKKGSDNAAKAQLQIARETNANNYRIAQENNAFNERMVDKM 67
S L G I+ GS++ GLF GS AAK QLQ RETN N IA +NN FNERM +
Sbjct 22 SALGAGLIAGAGSLINGLFSSNGSKQAAKYQLQAVRETNQANREIADQNNKFNERMWNLQ 81
Query 68 NDWNSAKNQRARLEEAGLNPYLMLdggsagtattaptadtssVQSAPDVGSTIasgaqql 127
N++N QRARLE AGLNPYLM+DGGSAG A +APTADTS Q APD+G+TIA G Q +
Sbjct 82 NEYNRPDMQRARLEAAGLNPYLMMDGGSAGIAESAPTADTSGTQIAPDIGNTIAGGYQAM 141
Query 128 gssissaasqisqqVYNSSLQEANVRKANADASSSEQDALLKGIESQFAAQRFLLDLKLK 187
G+SISSAASQI+Q + LQ+ANV K A+A ++ + ++FA FL++L+LK
Sbjct 142 GNSISSAASQIAQMTFQDDLQKANVAKTVAEAKNAHLQNQFDELRNEFAVANFLVNLRLK 201
Query 188 EMQGKVSEQDYYYLRDSMQDRLDSVKFQNTLTGSQSSYYNQMAGLVDVQRQIEKTNLDWL 247
+ QG +S+ + YLRDSMQDRLDSVKFQNTL+GSQSSYY+QMAGL DVQRQIE+TNLDWL
Sbjct 202 QKQGDISDYEANYLRDSMQDRLDSVKFQNTLSGSQSSYYSQMAGLTDVQRQIEQTNLDWL 261
Query 248 PREKQAGLAATLQNVRTMVSQMHLnyaqaknayamaalnyaqeaGVRLNNKLQDSIFDLS 307
P+EKQAGLAATLQN+RTMVS+M LNYAQAKNA+AMA+LNYA E G+R++N+L++S FDLS
Sbjct 262 PQEKQAGLAATLQNIRTMVSEMGLNYAQAKNAFAMASLNYANEEGLRIDNRLKESTFDLS 321
Query 308 VGMAEN 313
V +A+N
Sbjct 322 VKLAKN 327
>gi|494610273|ref|WP_007368519.1| hypothetical protein [Prevotella multiformis]
gi|324988545|gb|EGC20508.1| hypothetical protein HMPREF9141_0987 [Prevotella multiformis
DSM 16608]
Length=437
Score = 80.1 bits (196), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 43/89 (48%), Positives = 56/89 (63%), Gaps = 5/89 (6%)
Query 5 FDPLSLLSPGISAIGSIVGGLFG----KKGSDNAAKAQLQIARETNANNYRIAQENNAFN 60
DP S++ IS S++GGLFG K D A + LQIARE N N Y++ QE NAFN
Sbjct 64 IDP-SIIGSAISGGVSLLGGLFGGHSNKTAQDRANETNLQIAREANQNQYQMFQEQNAFN 122
Query 61 ERMVDKMNDWNSAKNQRARLEEAGLNPYL 89
ERM ++MN +NS Q R +AG+NPY+
Sbjct 123 ERMWNQMNQYNSPAAQMQRYTDAGINPYI 151
>gi|565841291|ref|WP_023924572.1| hypothetical protein [Prevotella nigrescens]
gi|564729909|gb|ETD29853.1| hypothetical protein HMPREF1173_00035 [Prevotella nigrescens
CC14M]
Length=396
Score = 71.2 bits (173), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 33/77 (43%), Positives = 50/77 (65%), Gaps = 0/77 (0%)
Query 15 ISAIGSIVGGLFGKKGSDNAAKAQLQIARETNANNYRIAQENNAFNERMVDKMNDWNSAK 74
+S +++GG+ G +A L+IARETNA N+++ Q N FN++M+DK N++
Sbjct 11 VSGASNLLGGIINGFGQKSANSTNLRIARETNAANFQMMQYQNEFNQKMLDKQNEYALPI 70
Query 75 NQRARLEEAGLNPYLML 91
NQR R E+AG+NPY L
Sbjct 71 NQRKRFEDAGINPYFAL 87
>gi|647452992|ref|WP_025792810.1| hypothetical protein [Prevotella histicola]
Length=424
Score = 59.3 bits (142), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/54 (52%), Positives = 38/54 (70%), Gaps = 0/54 (0%)
Query 36 KAQLQIARETNANNYRIAQENNAFNERMVDKMNDWNSAKNQRARLEEAGLNPYL 89
+ LQIARETN NY++ QE+NAFNE+M + N +N+ Q R EAG+NPY+
Sbjct 31 QTNLQIARETNQMNYQLFQESNAFNEKMYHEANAYNTPSAQMQRYAEAGINPYI 84
>gi|547226428|ref|WP_021963491.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103380|emb|CCY83991.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=416
Score = 55.8 bits (133), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 41/112 (37%), Positives = 52/112 (46%), Gaps = 27/112 (24%)
Query 6 DPLSLLSPGISAIGSIVGGLFGKKGSDNAAKAQLQIARETNANNYRI------------- 52
DP + S I A S +G +FG K + K LQIA+ N N R+
Sbjct 2 DPF-VGSALIGAGASFLGNIFGSKSQSDTNKTNLQIAQMNNEYNERMFNKQLEYNQDMFN 60
Query 53 ---------AQENNAFNERM----VDKMNDWNSAKNQRARLEEAGLNPYLML 91
++ N FN RM + +NSAK QRARLE AGLNPYLM+
Sbjct 61 QQVEYDQKKMEQQNNFNARMQNEAIGAQQVYNSAKAQRARLEAAGLNPYLMM 112
>gi|494822881|ref|WP_007558289.1| hypothetical protein [Bacteroides plebeius]
gi|198272097|gb|EDY96366.1| hypothetical protein BACPLE_00802 [Bacteroides plebeius DSM 17135]
Length=344
Score = 52.4 bits (124), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/63 (46%), Positives = 41/63 (65%), Gaps = 11/63 (17%)
Query 40 QIARETNANNYRIAQENNAFN----ERMVDKM-------NDWNSAKNQRARLEEAGLNPY 88
+IA TN N +IAQ +N +N ER +++ N++NSA +QR RLEEAGLNPY
Sbjct 38 EIAESTNQANIQIAQMSNEYNREQLERQIEQEWDMWNAENEYNSASSQRKRLEEAGLNPY 97
Query 89 LML 91
+M+
Sbjct 98 MMM 100
>gi|490418711|ref|WP_004291034.1| hypothetical protein [Bacteroides eggerthii]
gi|217986638|gb|EEC52972.1| hypothetical protein BACEGG_02723 [Bacteroides eggerthii DSM
20697]
Length=368
Score = 50.8 bits (120), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 21/45 (47%), Positives = 32/45 (71%), Gaps = 0/45 (0%)
Query 47 ANNYRIAQENNAFNERMVDKMNDWNSAKNQRARLEEAGLNPYLML 91
AN +++ ++N A+ M +K N++N QRARLE AGLNPY+M+
Sbjct 76 ANAWKLYEDNKAYQTEMWNKQNEYNDPSAQRARLEAAGLNPYMMM 120
>gi|496521296|ref|WP_009229579.1| hypothetical protein [Prevotella sp. oral taxon 317]
gi|288330567|gb|EFC69151.1| hypothetical protein HMPREF0670_00474 [Prevotella sp. oral taxon
317 str. F0108]
Length=429
Score = 47.8 bits (112), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 30/88 (34%), Positives = 49/88 (56%), Gaps = 8/88 (9%)
Query 8 LSLLSPGISAIGSIVGGLFGKKGSDNAAKAQLQIARETNANNYRIAQENNAFNERMV--D 65
L+++ I A +IVGGL + G+ A + RE N ++AQ++ ++M D
Sbjct 19 LAIIPALIGAGAAIVGGLLSRSGAKKANEQNYDFQREMW--NKQVAQQDKVNAQQMAYQD 76
Query 66 KMN----DWNSAKNQRARLEEAGLNPYL 89
K+N +W++ N R R+E+AG NPYL
Sbjct 77 KVNAENREWSTESNVRQRIEDAGYNPYL 104
>gi|575094358|emb|CDL65740.1| unnamed protein product [uncultured bacterium]
Length=328
Score = 45.8 bits (107), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 26/82 (32%), Positives = 46/82 (56%), Gaps = 7/82 (9%)
Query 17 AIGSIV---GGLF----GKKGSDNAAKAQLQIARETNANNYRIAQENNAFNERMVDKMND 69
A+G+++ GL+ K DN +QIA+ N + R+ ++ A+N M +K+ D
Sbjct 2 AVGTMIDIGAGLYTAISNKNAVDNTNSTNMQIAQMNNEWSERMMEKQMAYNTEMWEKVAD 61
Query 70 WNSAKNQRARLEEAGLNPYLML 91
+NS N+ + +AG+NPY+ L
Sbjct 62 YNSLPNKMQQARDAGVNPYMAL 83
>gi|639237431|ref|WP_024568108.1| hypothetical protein [Elizabethkingia anophelis]
Length=287
Score = 44.3 bits (103), Expect = 0.071, Method: Compositional matrix adjust.
Identities = 30/83 (36%), Positives = 43/83 (52%), Gaps = 19/83 (23%)
Query 8 LSLLSPGISAIGSIVGGLFGKKGSDNAAKAQLQIARETNANNYRIAQENNAFNERMVDKM 67
+SLLS I +G I+ G N++N +IA+EN AF M ++
Sbjct 1 MSLLS-AIPVVGGIIEGAMN------------------NSSNKKIARENRAFALDMWNRN 41
Query 68 NDWNSAKNQRARLEEAGLNPYLM 90
N++N+ Q RL+EAGLNP LM
Sbjct 42 NEYNTPLAQMQRLKEAGLNPNLM 64
Lambda K H a alpha
0.314 0.128 0.350 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2257338701640