bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-12_CDS_annotation_glimmer3.pl_2_4
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
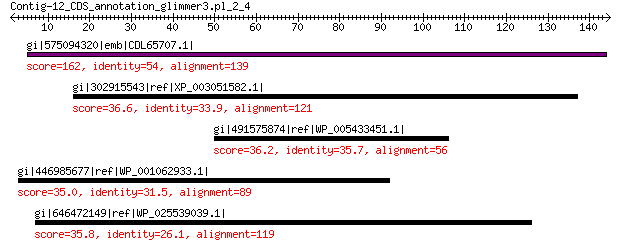
gi|575094320|emb|CDL65707.1| unnamed protein product 162 1e-47
gi|302915543|ref|XP_003051582.1| predicted protein 36.6 4.6
gi|491575874|ref|WP_005433451.1| protein disaggregation chaperone 36.2 5.8
gi|446985677|ref|WP_001062933.1| ribonuclease H 35.0 7.5
gi|646472149|ref|WP_025539039.1| hypothetical protein 35.8 8.5
>gi|575094320|emb|CDL65707.1| unnamed protein product [uncultured bacterium]
Length=148
Score = 162 bits (409), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 75/139 (54%), Positives = 103/139 (74%), Gaps = 0/139 (0%)
Query 5 EFLSYIPPIGEDLQHTKVSDDCTKLHSDIYLLHHIGDLNISNNLADAIKSRLQSVSDFMP 64
+ L YIPP+GE+LQH V + LHSD+Y+L + D+ +S N+ D I +RLQ V D+MP
Sbjct 7 DVLQYIPPLGEELQHQIVDKNTVCLHSDVYILTRLSDMKLSQNMVDIITNRLQEVKDYMP 66
Query 65 DDLRQSFDKLDDFQKMEVTDSRYAQWVSDKVSRTKQFMKEYDTALSKLKDSEETEKLKVA 124
D+LRQSFDKL+DF+KM++TDSRY QW+SD+V TK+F+ + D+ + K KDSE+ KLK A
Sbjct 67 DNLRQSFDKLNDFEKMDLTDSRYQQWLSDRVENTKEFISQLDSEIKKAKDSEDMAKLKQA 126
Query 125 QKNLRDFILRLGSAEESDN 143
LRDF+LRLGS + D+
Sbjct 127 NLALRDFVLRLGSPDIPDD 145
>gi|302915543|ref|XP_003051582.1| predicted protein [Nectria haematococca mpVI 77-13-4]
gi|256732521|gb|EEU45869.1| predicted protein [Nectria haematococca mpVI 77-13-4]
Length=1334
Score = 36.6 bits (83), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 41/148 (28%), Positives = 70/148 (47%), Gaps = 35/148 (24%)
Query 16 DLQHTKVSDDCTKLHSDIYLLHHIGDLNISNNLAD----------AIKSRLQS------- 58
+L+ K +D+ LH I LL H+G+++ISNNLA+ ++S L S
Sbjct 798 ELKEQKCADE---LHRTIALLSHLGEVHISNNLANYADDAHMKAMLMQSHLDSAAVSFNT 854
Query 59 ----VSDFMP-----DDLRQSFDKLDDFQKMEVTDSRYAQWVSDKVSRTKQFMKEYDTAL 109
V+ +P ++L Q F K + +T +R A+ ++ K R Q ++ +L
Sbjct 855 LRNLVNRVVPAEGEEEELAQHFAKKSEAV---ITQTRGAKVIAGKAVRALQDLRTR--SL 909
Query 110 SKLKDSEET-EKLKVAQKNLRDFILRLG 136
S L D+ E E+ A + L D ++G
Sbjct 910 SLLPDTTEAFEQCCEATQELADLARQIG 937
>gi|491575874|ref|WP_005433451.1| protein disaggregation chaperone [Sutterella wadsworthensis]
gi|404659231|gb|EKB32085.1| chaperone ClpB [Sutterella wadsworthensis 2_1_59BFAA]
Length=864
Score = 36.2 bits (82), Expect = 5.8, Method: Composition-based stats.
Identities = 20/56 (36%), Positives = 35/56 (63%), Gaps = 1/56 (2%)
Query 50 DAIKSRLQSVSDFMPDDLRQSFDKLDDFQKMEVTDSRYAQWVSDKVSRTKQFMKEY 105
DA K RLQ++ D + D L + + L++ K E +++ A+ V D++ RT+ M+EY
Sbjct 435 DASKKRLQAIEDEI-DKLSREYADLEEVWKKEKSEALGAKSVRDEIDRTRHQMEEY 489
>gi|446985677|ref|WP_001062933.1| ribonuclease H [Staphylococcus aureus]
gi|379021147|ref|YP_005297809.1| ribonuclease HI [Staphylococcus aureus subsp. aureus M013]
gi|545634849|ref|YP_008585419.1| hypothetical protein SA957_1323 [Staphylococcus aureus subsp.
aureus SA957]
gi|545637776|ref|YP_008587952.1| hypothetical protein SA40_1308 [Staphylococcus aureus subsp.
aureus SA40]
gi|359830456|gb|AEV78434.1| Ribonuclease HI [Staphylococcus aureus subsp. aureus M013]
gi|375369005|gb|EHS72897.1| ribonuclease HI [Staphylococcus aureus subsp. aureus IS-160]
gi|545581738|gb|AGW33820.1| hypothetical protein SA957_1323 [Staphylococcus aureus subsp.
aureus SA957]
gi|545584272|gb|AGW36353.1| hypothetical protein SA40_1308 [Staphylococcus aureus subsp.
aureus SA40]
gi|580636645|gb|EVG60329.1| ribonuclease HI [Staphylococcus aureus OCMM6067]
gi|580639742|gb|EVG63372.1| ribonuclease HI [Staphylococcus aureus OCMM6066]
gi|580787052|gb|EVI09277.1| ribonuclease HI [Staphylococcus aureus OCMM6095]
gi|582456274|gb|EVX49711.1| ribonuclease HI [Staphylococcus aureus F70893]
gi|582470175|gb|EVX63311.1| ribonuclease HI [Staphylococcus aureus F77047]
gi|582626752|gb|EVZ16816.1| ribonuclease HI [Staphylococcus aureus H48052]
gi|582629185|gb|EVZ19231.1| ribonuclease HI [Staphylococcus aureus H48054]
gi|582846644|gb|EWB32716.1| ribonuclease HI [Staphylococcus aureus W21932]
gi|585047054|gb|EWL69777.1| ribonuclease HI [Staphylococcus aureus F77917]
gi|587424380|gb|EWX05121.1| ribonuclease HI [Staphylococcus aureus H81433]
gi|587546137|gb|EWY25475.1| ribonuclease HI [Staphylococcus aureus W73738]
gi|593351165|gb|EXP42745.1| ribonuclease HI [Staphylococcus aureus W25797]
gi|599589146|gb|EYF20882.1| ribonuclease HI [Staphylococcus aureus F19490]
gi|599818775|gb|EYH43947.1| ribonuclease HI [Staphylococcus aureus W25799]
gi|600104072|gb|EYK08308.1| ribonuclease HI [Staphylococcus aureus M17033]
gi|600293261|gb|EYL93794.1| ribonuclease HI [Staphylococcus aureus M64056]
gi|600322284|gb|EYM22173.1| ribonuclease HI [Staphylococcus aureus T63897]
gi|600324715|gb|EYM24533.1| ribonuclease HI [Staphylococcus aureus T63898]
gi|616903029|gb|KAG94829.1| hypothetical protein W793_00732 [Staphylococcus aureus VET1518S]
gi|645299704|gb|KDP64360.1| ribonuclease HI [Staphylococcus aureus subsp. aureus 21321]
gi|670939939|gb|AII55894.1| hypothetical protein SA268_1327 [Staphylococcus aureus subsp.
aureus SA268]
Length=133
Score = 35.0 bits (79), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 28/97 (29%), Positives = 45/97 (46%), Gaps = 21/97 (22%)
Query 3 DNEFLSYIPPIGEDLQHTKVSDDCTKLHSDIYLLHHIGDLNISNNL--------ADAIKS 54
D + +YI +GE HT C IY L H +LN+SN L AD+I++
Sbjct 26 DEQHYTYIHELGEMDNHTAEWAAC------IYALEHARELNVSNALLYTDSKLIADSIEA 79
Query 55 RLQSVSDFMPDDLRQSFDKLDDFQKMEVTDSRYAQWV 91
++F P FD+++ F+K D + +W+
Sbjct 80 GYVKNANFKP-----YFDQIEIFEKS--FDLLFVKWI 109
>gi|646472149|ref|WP_025539039.1| hypothetical protein [Cytophagaceae bacterium JGI 0001001-B3]
Length=682
Score = 35.8 bits (81), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 31/123 (25%), Positives = 60/123 (49%), Gaps = 16/123 (13%)
Query 7 LSYIPPIGEDLQHTKVSDDCTKLHSDIYLLHHIGDL----NISNNLADAIKSRLQSVSDF 62
LS++ E + H V + C L + H+ GDL I N L+D I++ +
Sbjct 472 LSHVEKYFESISH--VDNLCVALSLQYEVFHYKGDLVAAEKIINKLSDIIET-------Y 522
Query 63 MPDDLRQSFDKLDDFQKMEVTDSRYAQWVSDKVSRTKQFMKEYDTALSKLKDSEETEKLK 122
DL+Q FD L + T ++A++ +D S++ + KEY+ + ++K ++ ++ +
Sbjct 523 EYKDLKQKFDSL---KSGGTTHEKFAKFYNDLASQSNEKQKEYEDMVREMKGWDQLDRTR 579
Query 123 VAQ 125
Q
Sbjct 580 ENQ 582
Lambda K H a alpha
0.315 0.131 0.365 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 441723247020