bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-12_CDS_annotation_glimmer3.pl_2_6
Length=254
Score E
Sequences producing significant alignments: (Bits) Value
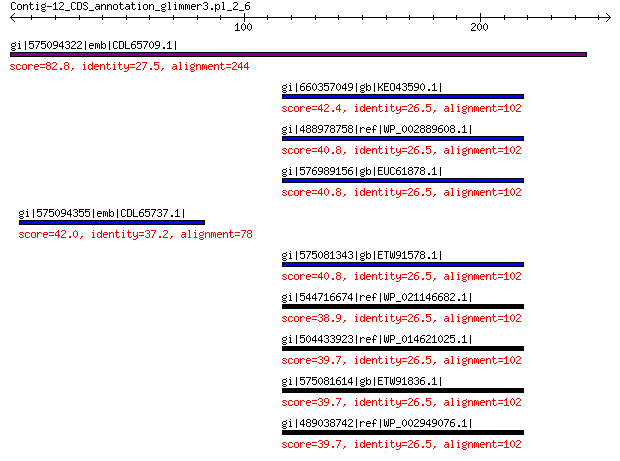
gi|575094322|emb|CDL65709.1| unnamed protein product 82.8 9e-15
gi|660357049|gb|KEO43590.1| NADPH-dependent FMN reductase 42.4 0.10
gi|488978758|ref|WP_002889608.1| oxidoreductase 40.8 0.16
gi|576989156|gb|EUC61878.1| flavin reductase 40.8 0.30
gi|575094355|emb|CDL65737.1| unnamed protein product 42.0 0.32
gi|575081343|gb|ETW91578.1| NADPH-dependent FMN reductase 40.8 0.39
gi|544716674|ref|WP_021146682.1| hypothetical protein 38.9 0.74
gi|504433923|ref|WP_014621025.1| NADPH-dependent FMN reductase 39.7 0.76
gi|575081614|gb|ETW91836.1| NADPH-dependent FMN reductase 39.7 0.78
gi|489038742|ref|WP_002949076.1| hypothetical protein 39.7 0.78
>gi|575094322|emb|CDL65709.1| unnamed protein product [uncultured bacterium]
Length=499
Score = 82.8 bits (203), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 67/247 (27%), Positives = 116/247 (47%), Gaps = 14/247 (6%)
Query 1 LSALSRPHSLHSRFFGQTLAEEEIQRAIVSQDFEYFRVHYRYSSKGYQIPYALWRSYYSR 60
L LS + HS GQ L+E+ I AI DF +F + + G Y++WRSYYSR
Sbjct 253 LVLLSNQKAYHSIQLGQILSEQSIVSAIQKGDFSFFERQFYLDTFGAANSYSVWRSYYSR 312
Query 61 FFPVFTGLVNMSNEKIFRLFEYWEEIGKLSGYYRVSNQVQWLKNWYYYYYSRPLETIPSP 120
FFP FT ++ E+ +R+ +E + L V + L +Y+Y++ P
Sbjct 313 FFPKFTCSSQLTYEQTYRVLTCYETLRDLFDTDSVGVICRRL--FYHYHFGYP------D 364
Query 121 IADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIWSNFYK 180
D+ L+ ++ + FSAL + AS+ F + ++ YF + +FY+
Sbjct 365 YHDIFDFLRFAYNAVLNSKDISLFSALRSCVSASRTFLRAAAMCGLTPTAYFRKYKDFYR 424
Query 181 YINLSLYKEHYLALENDSKYFRGWFD---RRFITPTAVLEFPLADSDFTNFVENSVSTHD 237
Y++LS + H+ S+Y +++ R I + + +DS+F+ F + +
Sbjct 425 YLDLSHLRSHFENCIASSEYSNNYYNIYQLRTINGNYIYD---SDSEFSRFRVSEQIRFE 481
Query 238 SLIKHRE 244
IKHR+
Sbjct 482 RSIKHRD 488
>gi|660357049|gb|KEO43590.1| NADPH-dependent FMN reductase [Streptococcus salivarius]
gi|660359472|gb|KEO45927.1| NADPH-dependent FMN reductase [Streptococcus salivarius]
Length=182
Score = 42.4 bits (98), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 43/102 (42%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+++
Sbjct 81 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KIVTVSSVANGGHNQLFDVY 130
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
+I + + ND+ W D +F+ VLE
Sbjct 131 KELLPFIRTQVVGDFTATCVNDT----AWVDGKFVATAEVLE 168
>gi|488978758|ref|WP_002889608.1| oxidoreductase [Streptococcus salivarius]
gi|400181892|gb|EJO16154.1| Oxidoreductase [Streptococcus salivarius K12]
Length=117
Score = 40.8 bits (94), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 43/102 (42%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+++
Sbjct 16 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KIVTVSSVANGGHNQLFDVY 65
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
+I + + ND+ W D +F+ VLE
Sbjct 66 KELLPFIRTQVVGDFTATRVNDT----AWVDGKFLATAEVLE 103
>gi|576989156|gb|EUC61878.1| flavin reductase [Streptococcus sp. ACS2]
Length=181
Score = 40.8 bits (94), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 44/102 (43%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+++
Sbjct 80 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KIVTVSSVANGGHNQLFDVY 129
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
+ +I + + ND+ W D +F+ VLE
Sbjct 130 KDLLPFIRTQVVGDFTATRVNDT----AWADGKFLATEEVLE 167
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 42.0 bits (97), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 40/80 (50%), Gaps = 8/80 (10%)
Query 5 SRPHSLHSRFFGQTLAEEEIQRAIVS--QDFEYFRVHYRYSSKGYQIPYALWRSYYSRFF 62
+RP SLHSRF GQ E ++ + +DF V S+K + LWRS YS F+
Sbjct 255 ARPFSLHSRFLGQGFLAHECEKVYETPVRDFVKRSVELNGSNKDFN----LWRSCYSVFY 310
Query 63 PVFTGLVNMSNEKIFRLFEY 82
P G S+ + RL+ Y
Sbjct 311 PKCKGFTRKSSSE--RLYTY 328
>gi|575081343|gb|ETW91578.1| NADPH-dependent FMN reductase [Streptococcus thermophilus MTH17CL396]
Length=184
Score = 40.8 bits (94), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 43/102 (42%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+ +
Sbjct 81 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KIVTVSSVANGGHNQLFDAY 130
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
++I + + ND+ W D +F+ VLE
Sbjct 131 KELLQFIRTQVVGDFTATRVNDT----AWVDGKFVVTEEVLE 168
>gi|544716674|ref|WP_021146682.1| hypothetical protein [Streptococcus sp. HSISS4]
gi|530811773|gb|EQC71520.1| hypothetical protein HSISS4_2043 [Streptococcus sp. HSISS4]
Length=125
Score = 38.9 bits (89), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 43/102 (42%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+ +
Sbjct 24 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KIVTVSSVANGGHNQLFDAY 73
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
+ +I + + ND+ W D +F+ VLE
Sbjct 74 KDLLPFIRTQVVGDFTATRVNDT----AWADGKFLATEEVLE 111
>gi|504433923|ref|WP_014621025.1| NADPH-dependent FMN reductase [Streptococcus thermophilus]
gi|386343677|ref|YP_006039841.1| hypothetical protein STH8232_0109 [Streptococcus thermophilus
JIM 8232]
gi|339277138|emb|CCC18886.1| hypothetical protein STH8232_0109 [Streptococcus thermophilus
JIM 8232]
Length=181
Score = 39.7 bits (91), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 42/102 (41%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+ +
Sbjct 80 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KIVTVSSVANGGHNQLFDAY 129
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
+I + + ND+ W D +F+ VLE
Sbjct 130 KELLPFIRTQVVGDFTATRVNDT----AWVDGKFVVTEEVLE 167
>gi|575081614|gb|ETW91836.1| NADPH-dependent FMN reductase [Streptococcus thermophilus M17PTZA496]
gi|585229509|gb|EWM60760.1| NADPH-dependent FMN reductase [Streptococcus thermophilus TH982]
Length=184
Score = 39.7 bits (91), Expect = 0.78, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 42/102 (41%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+ +
Sbjct 81 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KIVTVSSVANGGHNQLFDAY 130
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
+I + + ND+ W D +F+ VLE
Sbjct 131 KELLPFIRTQVVGDFTATRVNDT----AWVDGKFVVTEEVLE 168
>gi|489038742|ref|WP_002949076.1| hypothetical protein [Streptococcus thermophilus]
gi|444751579|gb|ELW76301.1| hypothetical protein IQ7_00379 [Streptococcus thermophilus MTCC
5461]
gi|444751643|gb|ELW76361.1| hypothetical protein IQ5_00353 [Streptococcus thermophilus MTCC
5460]
Length=183
Score = 39.7 bits (91), Expect = 0.78, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 42/102 (41%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+ +
Sbjct 80 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KMVTVSSVANGGHNQLFDAY 129
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
+I + + ND+ W D +F+ VLE
Sbjct 130 KELLPFIRTQVVGDFTATRVNDT----AWVDGKFVVTEEVLE 167
Lambda K H a alpha
0.325 0.138 0.449 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1155625517970