bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-13_CDS_annotation_glimmer3.pl_2_1
Length=298
Score E
Sequences producing significant alignments: (Bits) Value
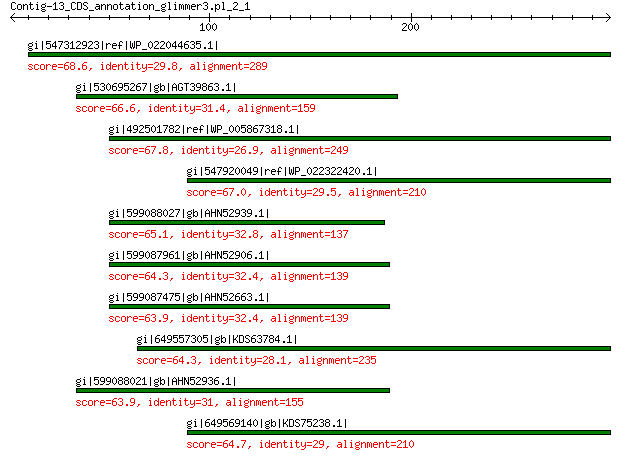
gi|547312923|ref|WP_022044635.1| putative uncharacterized protein 68.6 6e-10
gi|530695267|gb|AGT39863.1| major capsid protein 66.6 1e-09
gi|492501782|ref|WP_005867318.1| hypothetical protein 67.8 2e-09
gi|547920049|ref|WP_022322420.1| capsid protein VP1 67.0 3e-09
gi|599088027|gb|AHN52939.1| major capsid protein 65.1 3e-09
gi|599087961|gb|AHN52906.1| major capsid protein 64.3 5e-09
gi|599087475|gb|AHN52663.1| major capsid protein 63.9 6e-09
gi|649557305|gb|KDS63784.1| capsid family protein 64.3 7e-09
gi|599088021|gb|AHN52936.1| major capsid protein 63.9 7e-09
gi|649569140|gb|KDS75238.1| capsid family protein 64.7 2e-08
>gi|547312923|ref|WP_022044635.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208404|emb|CCZ76639.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=338
Score = 68.6 bits (166), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 86/334 (26%), Positives = 138/334 (41%), Gaps = 53/334 (16%)
Query 10 GLAVKTYQSDIFNNWIQTEWLEGENG---INAITAVDTSIG---SFTMDTLNLAKKVYNM 63
GL Y D+F N I+ +G + I + A+D +I S + L L K+ N
Sbjct 11 GLLSVPYSPDLFGNIIK----QGSSPAVEIEVMNALDLNISTGFSVAVPELRLRTKIQNW 66
Query 64 LNRIAVSDG----TYKSWMETVFSAKYVERTETPIYYGGMSQEII----FEEVVSTSATG 115
++R+ VS G +++ T SA YV + + + G+ Q I + + SA+G
Sbjct 67 MDRLFVSGGRVGDVFRTLWGTKSSAIYVNKPD----FLGVWQASINPSNVRAMANGSASG 122
Query 116 EEP-LGTLAGK-GRLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTMNWETMN 173
E+ LG LA R + EP + I + P YSQG + + +
Sbjct 123 EDANLGQLAACVDRYCDFSGHSGIDYYAKEPGTFMLITMLVPEPAYSQGLHPDLASISFG 182
Query 174 DLHKPALDMIGYQ-----------------DLTMEKAAWWTEEHTSD-TEFAQKSIGKTV 215
D P L+ IG+Q L E + W+ T + S+G+ V
Sbjct 183 DDFNPELNGIGFQLVPRHRFSMMPRGFNFTGLDQEASPWFGHTGTGVLVDPNMVSVGEEV 242
Query 216 AWVDYMTNYNKNYGNFASGENENFMTLDRNYNVENPD----------FT-TYIDPAKYNG 264
AW T+Y++ +G+FA N + L R + PD +T TYI+P +
Sbjct 243 AWSWLRTDYSRLHGDFAQNGNYQYWVLTRRFTTYFPDDGTGFYQDGEYTGTYINPLDWQY 302
Query 265 IFADQSRSSMNFWVQIGVNWKVRRKISAKSIPNL 298
+F DQ+ + NF + V +SA +P L
Sbjct 303 VFVDQTLMAGNFAYYGTFDLNVTSSLSANYMPYL 336
>gi|530695267|gb|AGT39863.1| major capsid protein, partial [uncultured marine Microviridae]
Length=263
Score = 66.6 bits (161), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 50/160 (31%), Positives = 71/160 (44%), Gaps = 4/160 (3%)
Query 34 NGINAITAVDTSIGSFTMDTLNLAKKVYNMLNRIAVSDGTYKSWMETVFSAKYVE-RTET 92
NG AI AV +S G ++TL A V +L R A Y +++ F + R +
Sbjct 106 NGYPAIYAVLSSTGGIPINTLRQAWMVQALLERDARGGTRYIEIIKSHFGVTSPDFRLQR 165
Query 93 PIYYGGMSQEIIFEEVVSTSATGEEPLGTLAGKGRLAPNKKGGKVVIKIDEPSYIIGICS 152
P Y GG S ++ + T G PLG + G G A + + E YIIGI S
Sbjct 166 PEYIGGGSTDLNITPIAQTVPGGGNPLGQIGGAGTAAGSHRASYAAT---EHGYIIGIIS 222
Query 153 ITPRLDYSQGNNFTMNWETMNDLHKPALDMIGYQDLTMEK 192
+ L Y QG N + T D + PA +G Q +T +
Sbjct 223 VKSELSYQQGINKMWDRHTRYDFYFPATAQLGEQAITQRE 262
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 67.8 bits (164), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 67/252 (27%), Positives = 109/252 (43%), Gaps = 13/252 (5%)
Query 50 TMDTLNLAKKVYNMLNRIAVSDGTYKSWMETVFSAKYVE-RTETPIYYGGMSQEIIFEEV 108
+++ L + + R A S Y + + F + + R + P + GG I EV
Sbjct 297 SINDLRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEV 356
Query 109 VSTSATGE-EPLGTLAGKGRLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTM 167
+ TSAT P +AG G A G K +E YIIGI SI PR Y QG
Sbjct 357 LQTSATDSTSPQANMAGHGISAGVNHGFKRYF--EEHGYIIGIMSIRPRTGYQQGVPKDF 414
Query 168 NWETMNDLHKPALDMIGYQDLTMEKAAWWTEEHTSDTEFAQKSIGKTVAWVDYMTNYNKN 227
D + P +G Q++ E+ +++ F G T + +Y + N+
Sbjct 415 RKFDNMDFYFPEFAHLGEQEIKNEEVYLQQTPASNNGTF-----GYTPRYAEYKYSMNEV 469
Query 228 YGNFASGENENFMTLDRNYNVENPDF-TTYIDPAKYNGIFADQSRSSMNFWVQIGVNWKV 286
+G+F N F L+R ++ E+P+ TT+++ N +FA S +W+Q+ + K
Sbjct 470 HGDFRG--NMAFWHLNRIFS-ESPNLNTTFVECNPSNRVFATAETSDDKYWIQLYQDVKA 526
Query 287 RRKISAKSIPNL 298
R + P L
Sbjct 527 LRLMPKYGTPML 538
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 67.0 bits (162), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 62/214 (29%), Positives = 99/214 (46%), Gaps = 16/214 (7%)
Query 89 RTETPIYYGGMSQEIIFEEVVSTSATGE-EPLGTLAGKGRLAPNKKGGKVVIKIDEPSYI 147
R + P + GG I EV+ TS+T E P +AG G A G K +E YI
Sbjct 352 RLQRPQFLGGGRMPISVSEVLQTSSTDETSPQANMAGHGISAGINNGFKHYF--EEHGYI 409
Query 148 IGICSITPRLDYSQG--NNFTMNWETMNDLHKPALDMIGYQDLTMEKAAWWTEEHTSDTE 205
IGI SITPR Y QG +FT ++ M D + P + Q++ ++ + D
Sbjct 410 IGIMSITPRSGYQQGVPRDFT-KFDNM-DFYFPEFAHLSEQEIKNQELFV-----SEDAA 462
Query 206 FAQKSIGKTVAWVDYMTNYNKNYGNFASGENENFMTLDRNYNVENPDF-TTYIDPAKYNG 264
+ + G T + +Y + ++ +G+F N +F L+R + + P+ TT+++ N
Sbjct 463 YNNGTFGYTPRYAEYKYHPSEAHGDFRG--NLSFWHLNRIFE-DKPNLNTTFVECKPSNR 519
Query 265 IFADQSRSSMNFWVQIGVNWKVRRKISAKSIPNL 298
+FA FWVQ+ + K R + P L
Sbjct 520 VFATSETEDDKFWVQMYQDVKALRLMPKYGTPML 553
>gi|599088027|gb|AHN52939.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=219
Score = 65.1 bits (157), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 45/137 (33%), Positives = 62/137 (45%), Gaps = 2/137 (1%)
Query 50 TMDTLNLAKKVYNMLNRIAVSDGTYKSWMETVFSAKYVERTETPIYYGGMSQEIIFEEVV 109
T++ L A ++ +L R A S Y ++ F +++ T P + GG S I V
Sbjct 77 TINQLRQAFQIQKLLERDARSGTRYAEIVKAHFGVNFMDVTYRPEFLGGTSTPINVTSVP 136
Query 110 STSATGEEPLGTLAGKGRLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTMNW 169
TS +G P GTLA G N GG E ++GI S+ L Y QG N +
Sbjct 137 QTSESGTTPQGTLAAFGTATVN--GGGFTKSFTEHCIVMGIASVRADLTYQQGLNRMFSR 194
Query 170 ETMNDLHKPALDMIGYQ 186
T D + PAL IG Q
Sbjct 195 STRYDFYFPALAHIGEQ 211
>gi|599087961|gb|AHN52906.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=210
Score = 64.3 bits (155), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 45/139 (32%), Positives = 63/139 (45%), Gaps = 2/139 (1%)
Query 50 TMDTLNLAKKVYNMLNRIAVSDGTYKSWMETVFSAKYVERTETPIYYGGMSQEIIFEEVV 109
T++ L A ++ +L R A S Y ++ F +++ T P + GG S I V
Sbjct 68 TINQLRQAFQIQKLLERDARSGTRYSEIVKAHFGVNFMDVTYRPEFLGGTSTPINVTSVP 127
Query 110 STSATGEEPLGTLAGKGRLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTMNW 169
TS +G P GTLA G N GG E ++GI S+ L Y QG N +
Sbjct 128 QTSESGTTPQGTLAAFGTATIN--GGGFTKSFTEHCIVMGIASVRADLTYQQGLNRMFSR 185
Query 170 ETMNDLHKPALDMIGYQDL 188
T D + PAL IG Q +
Sbjct 186 STRYDFYFPALAHIGEQSV 204
>gi|599087475|gb|AHN52663.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=210
Score = 63.9 bits (154), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 45/139 (32%), Positives = 63/139 (45%), Gaps = 2/139 (1%)
Query 50 TMDTLNLAKKVYNMLNRIAVSDGTYKSWMETVFSAKYVERTETPIYYGGMSQEIIFEEVV 109
T++ L A ++ +L R A S Y ++ F +++ T P + GG S I V
Sbjct 68 TINQLRQAFQIQKLLERDARSGTRYSEIVKAHFGVNFMDVTYRPEFLGGTSTPINVTSVP 127
Query 110 STSATGEEPLGTLAGKGRLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTMNW 169
TS +G P GTLA G N GG E ++GI S+ L Y QG N +
Sbjct 128 QTSESGTTPQGTLAAFGTATIN--GGGFTKSFTEHCILMGIASVRADLTYQQGLNRMFSR 185
Query 170 ETMNDLHKPALDMIGYQDL 188
T D + PAL IG Q +
Sbjct 186 STRYDFYFPALAHIGEQSV 204
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 64.3 bits (155), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 66/239 (28%), Positives = 104/239 (44%), Gaps = 15/239 (6%)
Query 64 LNRIAVSDGTYKSWMETVFSAKYVE-RTETPIYYGGMSQEIIFEEVVSTSATGE-EPLGT 121
R A S Y + + F + + R + P + GG I EV+ TS+T P
Sbjct 18 FERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQAN 77
Query 122 LAGKGRLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTMNWETMNDLHKPALD 181
+AG G A G +E YI+GI SI PR Y QG D + P
Sbjct 78 MAGHGISAGVNHG--FTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFA 135
Query 182 MIGYQDLTMEKAAWWTEEHTSDTEFA-QKSIGKTVAWVDYMTNYNKNYGNFASGENENFM 240
+G Q++ E E + ++++ A + + G T + +Y + N+ +G+F N F
Sbjct 136 HLGEQEIKNE------ELYLNESDAANEGTFGYTPRYAEYKYSQNEVHGDFRG--NMAFW 187
Query 241 TLDRNYNVENPDF-TTYIDPAKYNGIFADQSRSSMNFWVQIGVNWKVRRKISAKSIPNL 298
L+R + E P+ TT+++ N +FA S +WVQI + K R + P L
Sbjct 188 HLNRIFK-EKPNLNTTFVECNPSNRVFATAETSDDKYWVQIYQDIKALRLMPKYGTPML 245
>gi|599088021|gb|AHN52936.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=220
Score = 63.9 bits (154), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 48/155 (31%), Positives = 70/155 (45%), Gaps = 2/155 (1%)
Query 34 NGINAITAVDTSIGSFTMDTLNLAKKVYNMLNRIAVSDGTYKSWMETVFSAKYVERTETP 93
N A+ A +S + T++ L A ++ +L R A S Y ++ F +++ T P
Sbjct 62 NANRALYADLSSATAATINQLRQAFQIQKLLERDARSGTRYSEIVKAHFGVNFMDVTYRP 121
Query 94 IYYGGMSQEIIFEEVVSTSATGEEPLGTLAGKGRLAPNKKGGKVVIKIDEPSYIIGICSI 153
+ GG S + V TS +G P GTLA G N GG E ++GI S+
Sbjct 122 EFLGGSSTPVNVTSVPQTSESGTTPQGTLAAFGTATIN--GGGFTKSFTEHCIVMGIASV 179
Query 154 TPRLDYSQGNNFTMNWETMNDLHKPALDMIGYQDL 188
L Y QG N + T D + PAL IG Q +
Sbjct 180 RADLTYQQGLNRMFSRSTRYDFYFPALAHIGEQSV 214
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 64.7 bits (156), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 61/213 (29%), Positives = 95/213 (45%), Gaps = 14/213 (7%)
Query 89 RTETPIYYGGMSQEIIFEEVVSTSATGE-EPLGTLAGKGRLAPNKKGGKVVIKIDEPSYI 147
R + P + GG I EV+ TS+T P +AG G A G +E YI
Sbjct 189 RLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQANMAGHGISAGVNHG--FTRYFEEHGYI 246
Query 148 IGICSITPRLDYSQGNNFTMNWETMNDLHKPALDMIGYQDLTMEKAAWWTEEHTSDTEFA 207
+GI SI PR Y QG D + P +G Q++ E E + ++++ A
Sbjct 247 MGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNE------ELYLNESDAA 300
Query 208 -QKSIGKTVAWVDYMTNYNKNYGNFASGENENFMTLDRNYNVENPDF-TTYIDPAKYNGI 265
+ + G T + +Y + N+ +G+F N F L+R + E P+ TT+++ N +
Sbjct 301 NEGTFGYTPRYAEYKYSQNEVHGDFRG--NMAFWHLNRIFK-EKPNLNTTFVECNPSNRV 357
Query 266 FADQSRSSMNFWVQIGVNWKVRRKISAKSIPNL 298
FA S +WVQI + K R + P L
Sbjct 358 FATAETSDDKYWVQIYQDIKALRLMPKYGTPML 390
Lambda K H a alpha
0.315 0.131 0.395 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1564156586088