bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-13_CDS_annotation_glimmer3.pl_2_2
Length=99
Score E
Sequences producing significant alignments: (Bits) Value
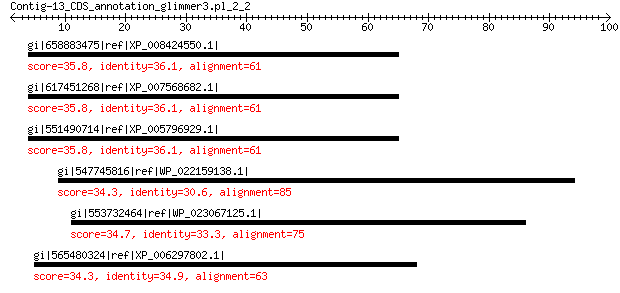
gi|658883475|ref|XP_008424550.1| PREDICTED: protocadherin-16-like 35.8 3.1
gi|617451268|ref|XP_007568682.1| PREDICTED: protocadherin-16-like 35.8 3.2
gi|551490714|ref|XP_005796929.1| PREDICTED: protocadherin-16-like 35.8 3.4
gi|547745816|ref|WP_022159138.1| putative uncharacterized protein 34.3 4.6
gi|553732464|ref|WP_023067125.1| phosphoribosylamine--glycine li... 34.7 5.8
gi|565480324|ref|XP_006297802.1| hypothetical protein CARUB_v100... 34.3 9.1
>gi|658883475|ref|XP_008424550.1| PREDICTED: protocadherin-16-like [Poecilia reticulata]
Length=3280
Score = 35.8 bits (81), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 22/65 (34%), Positives = 35/65 (54%), Gaps = 4/65 (6%)
Query 4 VEIKEGETIETK---VRRITEEKTPITDSAPIVYTNRTDGVIAGYNIRTDR-FEIALSAM 59
VE+KEG I T+ VR ++ E + +P++Y R DG AG+ I D + S++
Sbjct 1008 VELKEGTPINTRFLQVRAMSREVPGLGHLSPLMYHLRPDGDAAGFGIAADSGWLFVKSSL 1067
Query 60 DKVNK 64
D+ K
Sbjct 1068 DREQK 1072
>gi|617451268|ref|XP_007568682.1| PREDICTED: protocadherin-16-like [Poecilia formosa]
Length=3281
Score = 35.8 bits (81), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 22/65 (34%), Positives = 35/65 (54%), Gaps = 4/65 (6%)
Query 4 VEIKEGETIETK---VRRITEEKTPITDSAPIVYTNRTDGVIAGYNIRTDR-FEIALSAM 59
VE+KEG I T+ VR ++ E + +P++Y R DG AG+ I D + S++
Sbjct 1008 VELKEGTPINTRFLQVRAMSREVPGLGHLSPLMYHLRPDGDAAGFGIAADSGWLFVKSSL 1067
Query 60 DKVNK 64
D+ K
Sbjct 1068 DREQK 1072
>gi|551490714|ref|XP_005796929.1| PREDICTED: protocadherin-16-like [Xiphophorus maculatus]
Length=3269
Score = 35.8 bits (81), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 22/65 (34%), Positives = 35/65 (54%), Gaps = 4/65 (6%)
Query 4 VEIKEGETIETK---VRRITEEKTPITDSAPIVYTNRTDGVIAGYNIRTDR-FEIALSAM 59
VE+KEG I T+ VR ++ E + +P++Y R DG AG+ I D + S++
Sbjct 1008 VELKEGTPINTRFLQVRAMSREVPGLGHLSPLMYHLRPDGDAAGFGIAADSGWLFVKSSL 1067
Query 60 DKVNK 64
D+ K
Sbjct 1068 DREQK 1072
>gi|547745816|ref|WP_022159138.1| putative uncharacterized protein [Prevotella sp. CAG:520]
gi|524349928|emb|CDB05206.1| putative uncharacterized protein [Prevotella sp. CAG:520]
Length=133
Score = 34.3 bits (77), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 26/85 (31%), Positives = 43/85 (51%), Gaps = 4/85 (5%)
Query 9 GETIETKVRRITEEKTPITDSAPIVYTNRTDGVIAGYNIRTDRFEIALSAMDKVNKAKIA 68
E+IE + +R E T P++ RTD V+ N RT + S MD+++ AK+
Sbjct 19 HESIEKRAQREAREYTERNCPTPVLNYMRTDSVVFEMNTRT--YHYYCSIMDELDDAKVF 76
Query 69 TRNAKNKVEESDTNKEIESNTDTQA 93
N K V+ + K+I+ +T +A
Sbjct 77 EMNRKLIVD--NFMKQIDESTSLRA 99
>gi|553732464|ref|WP_023067125.1| phosphoribosylamine--glycine ligase [Lyngbya aestuarii]
gi|550815201|gb|ERT06617.1| phosphoribosylamine--glycine ligase [Lyngbya aestuarii BL J]
Length=423
Score = 34.7 bits (78), Expect = 5.8, Method: Composition-based stats.
Identities = 25/76 (33%), Positives = 37/76 (49%), Gaps = 3/76 (4%)
Query 11 TIETKVRRITEEKTPITDSAPIVYTNRTDGVIAGYNIRT-DRFEIALSAMDKVNKAKIAT 69
T +T V E +APIV + DG+ AG + DR E+A SA+D + + +
Sbjct 119 TAQTAVFTDAETAKAYVKTAPIVI--KADGLAAGKGVTVADRVELAHSAIDAIFAGEFGS 176
Query 70 RNAKNKVEESDTNKEI 85
N VEE T +E+
Sbjct 177 ENQTVVVEEYLTGQEV 192
>gi|565480324|ref|XP_006297802.1| hypothetical protein CARUB_v10013838mg [Capsella rubella]
gi|482566511|gb|EOA30700.1| hypothetical protein CARUB_v10013838mg [Capsella rubella]
Length=409
Score = 34.3 bits (77), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 33/64 (52%), Gaps = 1/64 (2%)
Query 5 EIKEGETIETKVRRITEEKTPITDSAPIVYTNR-TDGVIAGYNIRTDRFEIALSAMDKVN 63
EIKE E++ I E+ T + S P+VY NR TDG +A + + E S D++
Sbjct 90 EIKESGIDESETVNIAEDVTQLIGSTPMVYLNRVTDGCLADIAAKLESMEPCRSVKDRIG 149
Query 64 KAKI 67
+ I
Sbjct 150 LSMI 153
Lambda K H a alpha
0.304 0.122 0.312 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 441146700165