bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-19_CDS_annotation_glimmer3.pl_2_2
Length=167
Score E
Sequences producing significant alignments: (Bits) Value
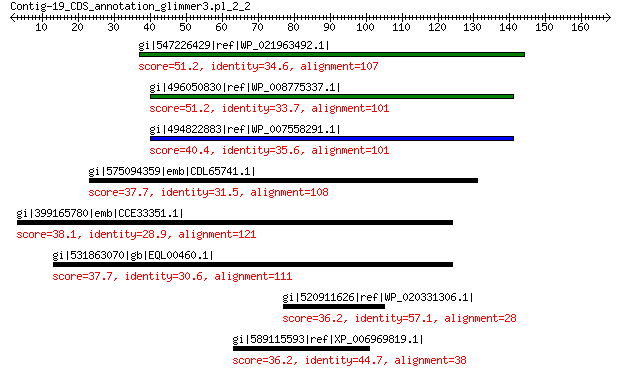
gi|547226429|ref|WP_021963492.1| putative uncharacterized protein 51.2 1e-05
gi|496050830|ref|WP_008775337.1| hypothetical protein 51.2 2e-05
gi|494822883|ref|WP_007558291.1| hypothetical protein 40.4 0.12
gi|575094359|emb|CDL65741.1| unnamed protein product 37.7 1.5
gi|399165780|emb|CCE33351.1| related to monooxigenase 38.1 1.9
gi|531863070|gb|EQL00460.1| ubiquinone biosynthesis monooxygenas... 37.7 2.8
gi|520911626|ref|WP_020331306.1| hypothetical protein 36.2 8.6
gi|589115593|ref|XP_006969819.1| predicted protein 36.2 9.6
>gi|547226429|ref|WP_021963492.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103381|emb|CCY83992.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=152
Score = 51.2 bits (121), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 37/121 (31%), Positives = 63/121 (52%), Gaps = 14/121 (12%)
Query 37 SGVLR----EQSPVEDFLFED--NFDGSR-----TFVSDYSVLSGQAAIDQMNPTQLRRY 85
SG+ R + SP+ +F+ E F+GS +V D +L Q +D + ++++
Sbjct 17 SGIERNNFVQPSPIREFMTEKVTYFNGSDKKTAIAYVDDIYMLFNQNRLDSVGRDTIQKW 76
Query 86 LNGLRPSS---SPYTHRYNDDFLLEYCKDRNIQSYTEMQAWLEHLISEGQSLENDVAAYQ 142
L+GL P S + D+ L+E CK R IQS +E+ AW E+L + S+ + + A +
Sbjct 77 LDGLTPRSDSLAKLRENVTDEQLMEICKSRYIQSSSELLAWSEYLNANYASILSSIEASK 136
Query 143 A 143
A
Sbjct 137 A 137
>gi|496050830|ref|WP_008775337.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448894|gb|EEO54685.1| hypothetical protein BSCG_01610 [Bacteroides sp. 2_2_4]
Length=154
Score = 51.2 bits (121), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 34/107 (32%), Positives = 57/107 (53%), Gaps = 9/107 (8%)
Query 40 LREQSPVEDFLFED---NFDGSRTFVSDYSVLSGQAAIDQMNPTQLRRYLNGL---RPSS 93
RE+SPV+ FLF++ + D S SD +L Q +D+++ T L Y N + P
Sbjct 29 FREESPVDQFLFQEVSVDGDTSIRLSSDIYMLFNQQRLDKLSQTSLLEYFNNISVTEPRF 88
Query 94 SPYTHRYNDDFLLEYCKDRNIQSYTEMQAWLEHLISEGQSLENDVAA 140
+ + D+ L+ + K R IQS +E+ AW +L++ S + +AA
Sbjct 89 NELRSKLGDEQLISFVKSRFIQSKSELMAWSNYLMN---STDEQIAA 132
>gi|494822883|ref|WP_007558291.1| hypothetical protein [Bacteroides plebeius]
gi|198272098|gb|EDY96367.1| hypothetical protein BACPLE_00803 [Bacteroides plebeius DSM 17135]
Length=140
Score = 40.4 bits (93), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 36/112 (32%), Positives = 54/112 (48%), Gaps = 11/112 (10%)
Query 40 LREQSPVEDFLFED-----NFDGSRTFVSDYSVLSGQAAIDQMNPTQLRRYLNGLRPSS- 93
LR SPVEDF E+ + T+ +D ++ Q +D M Q YL+ R +S
Sbjct 7 LRVPSPVEDFYREEVQTPLGSTPAVTYRNDIYMILNQRRLDSMTLAQFSDYLDHDRSASQ 66
Query 94 -SPYTHRYNDDFLLEYCKDRNIQSYTEMQAWLEHL----ISEGQSLENDVAA 140
S + +D+ L ++ K R IQ +E++AW +L E Q LE V A
Sbjct 67 LSQMREKMSDEQLHQFVKSRYIQHPSELRAWASYLDIEYSKEIQKLEEAVEA 118
>gi|575094359|emb|CDL65741.1| unnamed protein product [uncultured bacterium]
Length=156
Score = 37.7 bits (86), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 34/116 (29%), Positives = 54/116 (47%), Gaps = 8/116 (7%)
Query 23 NCTCRGVH-SAAVALSGVLREQSPVEDFLFEDNFDG----SRTFVSDYSVLSGQAAIDQM 77
N R +H S V + RE+SPVE+F + + S + V D +L Q +D+M
Sbjct 8 NAVSRKLHVSKVVDRLSIFREKSPVENFYIQKHKSKYGCESVSIVDDVYMLFNQQRLDRM 67
Query 78 NPTQLRRYLNGLRPSSSPYT---HRYNDDFLLEYCKDRNIQSYTEMQAWLEHLISE 130
L +L+ + S R +D L + K R IQS +E+ A+ +L S+
Sbjct 68 TNAALADWLSRTSRTDSQLASLRSRMSDVQLGAFIKSRYIQSRSELLAYSSYLESQ 123
>gi|399165780|emb|CCE33351.1| related to monooxigenase [Claviceps purpurea 20.1]
Length=496
Score = 38.1 bits (87), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 35/122 (29%), Positives = 54/122 (44%), Gaps = 15/122 (12%)
Query 3 KDVIIRGFAR-YSLNRDNQLHNCTCRGVHSAAVALSGVLREQSPVEDFLFEDNFDGSRTF 61
+ V R + R YS ++D ++++ C G A ++L LR SP+ L R
Sbjct 25 RSVTHRQWTRSYSTSKD-EIYDVVCIGGGPAGLSLLAALRA-SPITSRL--------RVA 74
Query 62 VSDYSVLSGQAAIDQMNPTQLRRYLNGLRPSSSPYTHRYNDDFLLEYCKDRNIQSYTEMQ 121
+ + LS AA + PTQ + L P+S+ Y H + K IQ + EMQ
Sbjct 75 LVEAQDLSKTAAF-ALPPTQFSNRCSSLTPTSAQYLHSIGA---WAHLKRERIQEFQEMQ 130
Query 122 AW 123
W
Sbjct 131 VW 132
>gi|531863070|gb|EQL00460.1| ubiquinone biosynthesis monooxygenase COQ6 [Ophiocordyceps sinensis
CO18]
Length=487
Score = 37.7 bits (86), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 34/111 (31%), Positives = 48/111 (43%), Gaps = 14/111 (13%)
Query 13 YSLNRDNQLHNCTCRGVHSAAVALSGVLREQSPVEDFLFEDNFDGSRTFVSDYSVLSGQA 72
YS+ D Q+++ C G A ++L LR SPV L R + + LS A
Sbjct 30 YSIKTD-QIYDVVCVGGGPAGLSLLAALRS-SPVTSQL--------RVALVEAQHLSKTA 79
Query 73 AIDQMNPTQLRRYLNGLRPSSSPYTHRYNDDFLLEYCKDRNIQSYTEMQAW 123
A + PTQ + L P+S+ + N + K IQ Y EMQ W
Sbjct 80 AFS-LPPTQFSNRCSSLTPASAQF---LNSIGAWSHMKRERIQEYHEMQVW 126
>gi|520911626|ref|WP_020331306.1| hypothetical protein [Vibrio fluvialis]
gi|519729296|gb|EPP20649.1| hypothetical protein L910_1765 [Vibrio fluvialis PG41]
Length=577
Score = 36.2 bits (82), Expect = 8.6, Method: Composition-based stats.
Identities = 16/28 (57%), Positives = 17/28 (61%), Gaps = 0/28 (0%)
Query 77 MNPTQLRRYLNGLRPSSSPYTHRYNDDF 104
+NP Q Y NGL S PY RYNDDF
Sbjct 538 LNPGQAFIYFNGLPISDKPYMMRYNDDF 565
>gi|589115593|ref|XP_006969819.1| predicted protein [Trichoderma reesei QM6a]
gi|340513944|gb|EGR44217.1| predicted protein [Trichoderma reesei QM6a]
gi|572273203|gb|ETR96875.1| fungalysin metallopeptidase [Trichoderma reesei RUT C-30]
Length=640
Score = 36.2 bits (82), Expect = 9.6, Method: Composition-based stats.
Identities = 17/38 (45%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 63 SDYSVLSGQAAIDQMNPTQLRRYLNGLRPSSSPYTHRY 100
++Y+ G AI Q+NPT YLN RPSS T Y
Sbjct 290 TNYTTTRGNNAIAQVNPTGGNDYLNNYRPSSPSRTFEY 327
Lambda K H a alpha
0.318 0.132 0.388 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 440304771465