bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-19_CDS_annotation_glimmer3.pl_2_3
Length=365
Score E
Sequences producing significant alignments: (Bits) Value
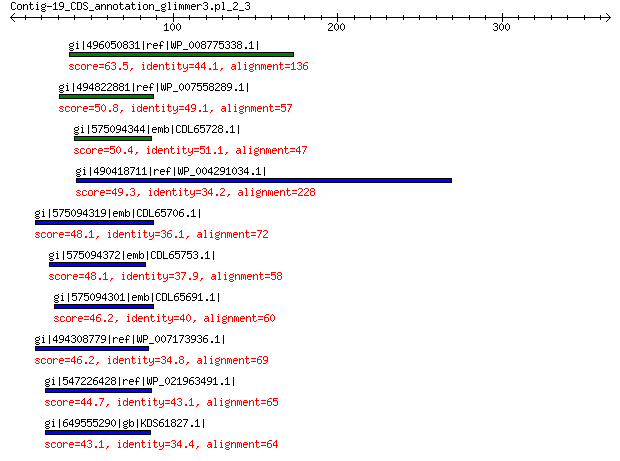
gi|496050831|ref|WP_008775338.1| predicted protein 63.5 6e-08
gi|494822881|ref|WP_007558289.1| hypothetical protein 50.8 7e-04
gi|575094344|emb|CDL65728.1| unnamed protein product 50.4 9e-04
gi|490418711|ref|WP_004291034.1| hypothetical protein 49.3 0.002
gi|575094319|emb|CDL65706.1| unnamed protein product 48.1 0.006
gi|575094372|emb|CDL65753.1| unnamed protein product 48.1 0.006
gi|575094301|emb|CDL65691.1| unnamed protein product 46.2 0.023
gi|494308779|ref|WP_007173936.1| hypothetical protein 46.2 0.026
gi|547226428|ref|WP_021963491.1| putative uncharacterized protein 44.7 0.081
gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 43.1 0.17
>gi|496050831|ref|WP_008775338.1| predicted protein [Bacteroides sp. 2_2_4]
gi|229448895|gb|EEO54686.1| hypothetical protein BSCG_01611 [Bacteroides sp. 2_2_4]
Length=381
Score = 63.5 bits (153), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 60/149 (40%), Positives = 86/149 (58%), Gaps = 20/149 (13%)
Query 37 QRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNPYLMMNggsagtass 96
Q++ ++W+ K A MF+ATNE+NSA QR R E AGLNPY+MMN GSAGTA++
Sbjct 66 QQVSDQWSFYNDAKQN--AWDMFNATNEYNSASAQRERYEAAGLNPYVMMNTGSAGTAAA 123
Query 97 tsantvsgasgsggtPYQYTPTNIIGDVASFAGAMKSLSEA----------RKTNTESDL 146
TSA + + + G TP +P + A ++G M+ L +A KT E+
Sbjct 124 TSATSATAPTKQGITPPTASPYS-----ADYSGIMQGLGQAIDQLSSIPDKAKTIAETGN 178
Query 147 L---GKYGDSDYSSRIANTEADTYFKQRQ 172
L GKY ++ +RIAN +ADT+ K+ Q
Sbjct 179 LKIEGKYKAAEAIARIANIKADTHSKKEQ 207
>gi|494822881|ref|WP_007558289.1| hypothetical protein [Bacteroides plebeius]
gi|198272097|gb|EDY96366.1| hypothetical protein BACPLE_00802 [Bacteroides plebeius DSM 17135]
Length=344
Score = 50.8 bits (120), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 28/57 (49%), Positives = 36/57 (63%), Gaps = 14/57 (25%)
Query 31 KHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNPYLMMN 87
+ QLE Q I+ EW M++A NE+NSA +QR RLEEAGLNPY+MM+
Sbjct 59 REQLERQ-IEQEW-------------DMWNAENEYNSASSQRKRLEEAGLNPYMMMD 101
>gi|575094344|emb|CDL65728.1| unnamed protein product [uncultured bacterium]
Length=368
Score = 50.4 bits (119), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 24/47 (51%), Positives = 30/47 (64%), Gaps = 0/47 (0%)
Query 40 QNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNPYLMM 86
Q EW S E+QK RDF M++ NE+N Q RLEEAG+NP+ M
Sbjct 49 QMEWQSQEAQKQRDFQLDMWNRNNEYNKPDEQMKRLEEAGINPWQSM 95
>gi|490418711|ref|WP_004291034.1| hypothetical protein [Bacteroides eggerthii]
gi|217986638|gb|EEC52972.1| hypothetical protein BACEGG_02723 [Bacteroides eggerthii DSM
20697]
Length=368
Score = 49.3 bits (116), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 78/250 (31%), Positives = 112/250 (45%), Gaps = 28/250 (11%)
Query 41 NEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNPYLMMNggsagtasstsan 100
N W E K+ + M++ NE+N QRARLE AGLNPY+MMNGGSAG A S S
Sbjct 77 NAWKLYEDNKA--YQTEMWNKQNEYNDPSAQRARLEAAGLNPYMMMNGGSAGVAGSVSGT 134
Query 101 tvsgasgsggtPYQYTPTNIIGDVASFAGAMKSLSEARKT----------NTESDLLGKY 150
S S + P A ++G M+ L A T N ++D L
Sbjct 135 QGSAPSAGSPSAQGVQPPTATPYSADYSGVMQGLGHAIDTIMTGSQRNIQNAQADNLRIE 194
Query 151 GDSDYSSRIANTEADTYFKQRQSDVATA-QRANLLLSSKAQEIMNMYLP----QEKQIEL 205
G S IA TY + + D A QR +LSS +++ + +QI+
Sbjct 195 GKYIASKAIAELY-KTYNEAKNDDERVAIQR---VLSSIQKDLSASQVAVNNENVRQIQA 250
Query 206 STLGAQYWNLIRDGSIK---EEQAKNLLATRLEIAARTAGQHISNKVAR----STADSII 258
T A NL+R+ +K EQ L +IA + A ++++ K AR A++I+
Sbjct 251 QTKIAVTENLLREQQLKFLPYEQRTQLALGAADIALKYAQKNLTEKQARHEIEKLAETIV 310
Query 259 DATNTAKMNE 268
A A N+
Sbjct 311 RANGQAMQNQ 320
>gi|575094319|emb|CDL65706.1| unnamed protein product [uncultured bacterium]
Length=396
Score = 48.1 bits (113), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 26/72 (36%), Positives = 43/72 (60%), Gaps = 0/72 (0%)
Query 16 VVNAIGNNRQGSKNRKHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARL 75
++N + ++ + K+QL+ R N+ + ++ F + M++ NE+N QRARL
Sbjct 35 LINGLFSSNGSKQAAKYQLQAVRETNQANREIADQNNKFNERMWNLQNEYNRPDMQRARL 94
Query 76 EEAGLNPYLMMN 87
E AGLNPYLMM+
Sbjct 95 EAAGLNPYLMMD 106
>gi|575094372|emb|CDL65753.1| unnamed protein product [uncultured bacterium]
Length=385
Score = 48.1 bits (113), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 22/58 (38%), Positives = 37/58 (64%), Gaps = 0/58 (0%)
Query 25 QGSKNRKHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNP 82
+ K R+ + +QNE+ +SE++K+R F KS+++ + WNS NQ + +AGLNP
Sbjct 55 EAEKARQFNSQQTALQNEFNASEAEKNRAFQKSLYERSLSWNSPSNQLKMMADAGLNP 112
>gi|575094301|emb|CDL65691.1| unnamed protein product [uncultured bacterium]
Length=437
Score = 46.2 bits (108), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 37/60 (62%), Gaps = 3/60 (5%)
Query 28 KNRKHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNPYLMMN 87
K +HQ E R + E+ RDFA+ M+ TN++N+ Q+ RLE+AG+NPY+ M+
Sbjct 42 KFERHQAEDAR---NFTHQENALQRDFARQMWKDTNDYNTPIAQKQRLEQAGMNPYVNMD 98
>gi|494308779|ref|WP_007173936.1| hypothetical protein [Prevotella bergensis]
gi|270333033|gb|EFA43819.1| hypothetical protein HMPREF0645_1833 [Prevotella bergensis DSM
17361]
Length=412
Score = 46.2 bits (108), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 24/69 (35%), Positives = 36/69 (52%), Gaps = 0/69 (0%)
Query 16 VVNAIGNNRQGSKNRKHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARL 75
V+ A G++ +NR+HQ +M Q + +Q+ DF + + WN N R R+
Sbjct 22 VLGAAGSSNLNRRNRRHQWDMMLQQQAYNDKVNQQQMDFQREVNQQNFAWNDPSNIRKRI 81
Query 76 EEAGLNPYL 84
E AG NPYL
Sbjct 82 EAAGYNPYL 90
>gi|547226428|ref|WP_021963491.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103380|emb|CCY83991.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=416
Score = 44.7 bits (104), Expect = 0.081, Method: Compositional matrix adjust.
Identities = 28/69 (41%), Positives = 38/69 (55%), Gaps = 4/69 (6%)
Query 22 NNRQGSKNRKHQLEMQRIQNEWASSESQKSRDFAKSM----FDATNEWNSAKNQRARLEE 77
N R +K ++ +M Q E+ + ++ +F M A +NSAK QRARLE
Sbjct 44 NERMFNKQLEYNQDMFNQQVEYDQKKMEQQNNFNARMQNEAIGAQQVYNSAKAQRARLEA 103
Query 78 AGLNPYLMM 86
AGLNPYLMM
Sbjct 104 AGLNPYLMM 112
>gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649557306|gb|KDS63785.1| hypothetical protein M095_3404 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649559158|gb|KDS65545.1| hypothetical protein M096_4689 [Parabacteroides distasonis str.
3999B T(B) 6]
gi|649560567|gb|KDS66875.1| hypothetical protein M095_2448 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561016|gb|KDS67303.1| hypothetical protein M095_2410 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649562727|gb|KDS68911.1| hypothetical protein M096_3341 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=288
Score = 43.1 bits (100), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 36/64 (56%), Gaps = 0/64 (0%)
Query 22 NNRQGSKNRKHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLN 81
NN+ K +E+ + Q +W E++K+ + M++ NE+NS Q AR+ AGLN
Sbjct 24 NNKAVQDTNKANMEIAKYQAQWQQQENEKAYQRSLKMWNLQNEYNSPTQQMARIRAAGLN 83
Query 82 PYLM 85
P L+
Sbjct 84 PNLV 87
Lambda K H a alpha
0.313 0.127 0.359 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2216296179792