bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-21_CDS_annotation_glimmer3.pl_2_2
Length=313
Score E
Sequences producing significant alignments: (Bits) Value
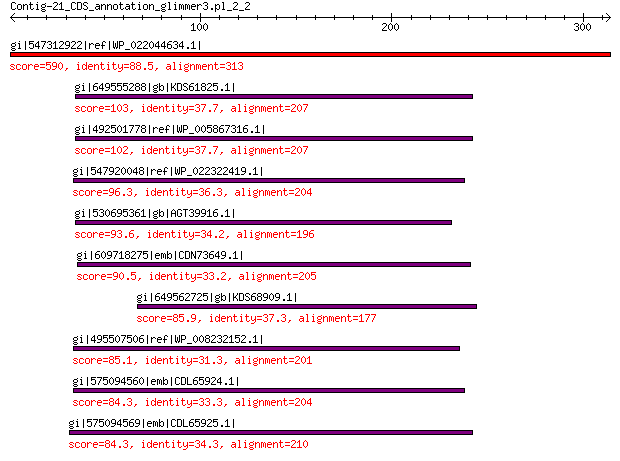
gi|547312922|ref|WP_022044634.1| putative replication initiation... 590 0.0
gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 103 4e-22
gi|492501778|ref|WP_005867316.1| hypothetical protein 102 9e-22
gi|547920048|ref|WP_022322419.1| putative replication protein 96.3 9e-20
gi|530695361|gb|AGT39916.1| replication initiator 93.6 8e-19
gi|609718275|emb|CDN73649.1| conserved hypothetical protein 90.5 7e-18
gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 85.9 3e-16
gi|495507506|ref|WP_008232152.1| hypothetical protein 85.1 7e-16
gi|575094560|emb|CDL65924.1| unnamed protein product 84.3 2e-15
gi|575094569|emb|CDL65925.1| unnamed protein product 84.3 2e-15
>gi|547312922|ref|WP_022044634.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
gi|524208442|emb|CCZ76638.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
Length=320
Score = 590 bits (1521), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 277/313 (88%), Positives = 295/313 (94%), Gaps = 0/313 (0%)
Query 1 MIVNRRYKDMTFNEVVDYAETYYGCFWPPDYYLEVPCGYCHSCQKSYNNQYRIRLLYELR 60
+IVNRRY +MT E+V+YA+ YYGCFWPPDY LEVPCGYCHSCQKSYNNQYRIRLLYELR
Sbjct 8 VIVNRRYANMTNTEIVNYAKVYYGCFWPPDYILEVPCGYCHSCQKSYNNQYRIRLLYELR 67
Query 61 KYPPGTCLFVTLTFDDDNLKKFSKDTNKAVRLFLDRLRKDYGKQIRHWFVCEFGTLYGRP 120
KYPPGTCLFVTLTF+DD+L+KFSKDTNKAVRLFLDR RK YGKQIRHWFVCEFGTL+GRP
Sbjct 68 KYPPGTCLFVTLTFNDDSLEKFSKDTNKAVRLFLDRFRKVYGKQIRHWFVCEFGTLHGRP 127
Query 121 HYHGILFDVPQTLIDGYSPDVPGHHPLLASRWKYGFVFVGYVSDETCSYITKYVTKSING 180
HYHGILF+VPQ LIDGY D+PGHHPLLAS WKYGFVFVGYVSDETCSYITKYVTKSING
Sbjct 128 HYHGILFNVPQALIDGYDSDMPGHHPLLASCWKYGFVFVGYVSDETCSYITKYVTKSING 187
Query 181 DKVRPRIISSFGIGSNYFDTEESTLHKLGGQRYQPFMVLNGFQQAMPRYYYNKIFSDVDK 240
DKVRPR+ISSFGIGSNY +TEES+LHKLG QRYQPFMVLNGFQQAMPRYYYNKIFSDVDK
Sbjct 188 DKVRPRVISSFGIGSNYLNTEESSLHKLGNQRYQPFMVLNGFQQAMPRYYYNKIFSDVDK 247
Query 241 QNIVLDRFVNPPVEFSWQGQKFSSKLERDEMRRSTLNQNITSGLTPALPLPHTERVSSFD 300
QN+V+DR +NPPVEFSWQGQKFSSKLERDEMRRSTLNQNI SGLTP LPLPHTERVSSFD
Sbjct 248 QNMVVDRLINPPVEFSWQGQKFSSKLERDEMRRSTLNQNIASGLTPVLPLPHTERVSSFD 307
Query 301 RFKENMDKNKEFK 313
FK+ MDKNKEFK
Sbjct 308 IFKKYMDKNKEFK 320
>gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649560564|gb|KDS66872.1| hypothetical protein M095_2449 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561011|gb|KDS67298.1| hypothetical protein M095_2409 [Parabacteroides distasonis str.
3999B T(B) 4]
Length=284
Score = 103 bits (256), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 78/230 (34%), Positives = 121/230 (53%), Gaps = 37/230 (16%)
Query 35 VPCGYCHSCQKSYNNQYRIRLLYELRKYPPGTCLFVTLTFDDDNL--KKFSKD------- 85
VPCG C +C+K+ + RL E +YP LFVTLT+DD+++ +D
Sbjct 15 VPCGRCVNCRKNKRQSWVYRLQAEADEYP--FSLFVTLTYDDEHIPTAMIGEDLFKTTVG 72
Query 86 --TNKAVRLFLDRLRKDYGK-QIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVP 142
+ + ++LF+ RLRK Y + ++R++ E+G+ GRPHYH ILF P T
Sbjct 73 VVSKRDIQLFMKRLRKKYAQYRLRYFLTSEYGSQGGRPHYHMILFGFPFT---------- 122
Query 143 GHH--PLLASRWKYGFVFVGYVSDETCSYITKYV-TKSINGDKVR------PRIISSF-- 191
G H LLA WK GFV ++ + SY+TKY+ KS+ D ++ P ++ S
Sbjct 123 GKHGGDLLAECWKNGFVQAHPLTTKEISYVTKYMYEKSMIPDILKGVKEYQPFMLCSKMP 182
Query 192 GIGSNYFDTEESTLHKLGGQRYQPFMVLNGFQQAMPRYYYNKIFSDVDKQ 241
GIG ++ + ++L + Y NG + AMPRYY +K++ D K+
Sbjct 183 GIGYHFLREQILDFYRLHPRDY--VRAFNGMRMAMPRYYADKLYDDDMKE 230
>gi|492501778|ref|WP_005867316.1| hypothetical protein [Parabacteroides distasonis]
gi|409230407|gb|EKN23271.1| hypothetical protein HMPREF1059_03256 [Parabacteroides distasonis
CL09T03C24]
Length=284
Score = 102 bits (254), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 78/230 (34%), Positives = 121/230 (53%), Gaps = 37/230 (16%)
Query 35 VPCGYCHSCQKSYNNQYRIRLLYELRKYPPGTCLFVTLTFDDDNL--KKFSKDTNKA--- 89
VPCG C +C+K+ + RL E +YP LFVTLT+DD+++ +D K+
Sbjct 15 VPCGRCVNCRKNKRQSWVYRLQAEADEYP--FSLFVTLTYDDEHMPTAMIGEDLFKSTVG 72
Query 90 ------VRLFLDRLRKDYGK-QIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVP 142
++LF+ RLRK Y + ++R++ E+G+ GRPHYH ILF P T
Sbjct 73 VVSKRDIQLFMKRLRKKYDQYRLRYFLTSEYGSQGGRPHYHMILFGFPFT---------- 122
Query 143 GHH--PLLASRWKYGFVFVGYVSDETCSYITKYV-TKSINGDKVR------PRIISSF-- 191
G H LLA WK GFV ++ + +Y+TKY+ KS+ D ++ P ++ S
Sbjct 123 GKHGGDLLAECWKNGFVQAHPLTTKEIAYVTKYMYEKSMVPDILKDVKEYQPFMLCSRIP 182
Query 192 GIGSNYFDTEESTLHKLGGQRYQPFMVLNGFQQAMPRYYYNKIFSDVDKQ 241
GIG ++ + ++L + Y NG + AMPRYY +K++ D K+
Sbjct 183 GIGYHFLREQILDFYRLHPRDY--VRAFNGMRMAMPRYYADKLYDDDMKE 230
>gi|547920048|ref|WP_022322419.1| putative replication protein [Parabacteroides merdae CAG:48]
gi|524592960|emb|CDD13572.1| putative replication protein [Parabacteroides merdae CAG:48]
Length=278
Score = 96.3 bits (238), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 74/225 (33%), Positives = 119/225 (53%), Gaps = 33/225 (15%)
Query 34 EVPCGYCHSCQKSYNNQYRIRLLYELRKYPPGTCLFVTLTFDDDNL--KKFSKD---TNK 88
+VPCG+C +C+++ + RL E ++YP LFVTLT+DD++L ++ D TN
Sbjct 9 KVPCGWCVNCRQNKRQSWVYRLQAEAKEYP--LSLFVTLTYDDEHLPIERIGSDLFQTNV 66
Query 89 AV------RLFLDRLRKDYGK-QIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDV 141
AV +LF+ RLRK Y ++R++ E+G GRPHYH ILF P ++ +
Sbjct 67 AVVSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGFP------FTGKM 120
Query 142 PGHHPLLASRWKYGFVFVGYVSDETCSYITKYV-TKSI------NGDKVRPRIISSF--G 192
G LLA W+ GFV ++ + +Y+ KY+ KS+ + K +P ++ S G
Sbjct 121 AGD--LLAECWQNGFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPG 178
Query 193 IGSNYFDTEESTLHKLGGQRYQPFMVLNGFQQAMPRYYYNKIFSD 237
IG + + ++ + Y G + AMPRYY +K++ D
Sbjct 179 IGFGFMKADIIEFYRRHPRDY--VRAWAGHKMAMPRYYADKLYDD 221
>gi|530695361|gb|AGT39916.1| replication initiator [Marine gokushovirus]
Length=289
Score = 93.6 bits (231), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 67/220 (30%), Positives = 101/220 (46%), Gaps = 28/220 (13%)
Query 35 VPCGYCHSCQKSYNNQYRIRLLYELRKYPPGTCLFVTLTFDDDNLKKFSKDT---NKAVR 91
+PCG C C+ Y+ Q+ IR ++E + + F+TLTFD++++ K N +
Sbjct 30 LPCGQCIGCRLDYSRQWAIRCVHEAQTHEDNC--FITLTFDNEHIAKRKNPESLDNTEFQ 87
Query 92 LFLDRLRKDYGKQIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVPGHHPL---- 147
F+ RLRK Y +IR + E+G RPHYH +LF D G L
Sbjct 88 RFMKRLRKKYPHKIRFFHCGEYGDQNKRPHYHALLFG--HDFKDKKLWSNKGDFKLFVSQ 145
Query 148 -LASRWKYGFVFVGYVSDETCSYITKYVTKSINGDKVRP---RIISSFGIGSNYFDTEES 203
LA W YGF +G VS +T +Y +YV K + GD + G N E
Sbjct 146 ELAELWPYGFHTIGAVSFDTAAYCARYVMKKVTGDAAASHYREVDLETGEVINEIKPEYC 205
Query 204 TLHKLGGQRYQ-------------PFMVLNGFQQAMPRYY 230
T+ ++ G Y+ ++V+NG++ PRYY
Sbjct 206 TMSRMPGIGYEWYQKYGYHDCHKHDYIVINGYKVRPPRYY 245
>gi|609718275|emb|CDN73649.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=265
Score = 90.5 bits (223), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 68/217 (31%), Positives = 102/217 (47%), Gaps = 29/217 (13%)
Query 36 PCGYCHSCQKSYNNQYRIRLLYELRKYPPGTCLFVTLTFDDDNL----KKFSKDTNKAVR 91
PCG C C+K+ N + RL EL+ + FVTLT+ D L + +
Sbjct 24 PCGKCLECRKARTNSWFARLTEELK--VSKSAHFVTLTYSDVYLPYSDNGLISLDYRDFQ 81
Query 92 LFLDRLRKDYGKQIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVPGHHPLLASR 151
LF+ R RK +I+++ V E+G RPHYH I+F V ID +
Sbjct 82 LFMKRARKLQKSKIKYFLVGEYGAQTYRPHYHAIVFGVEN--IDAF-----------LGE 128
Query 152 WKYGFVFVGYVSDETCSYITKYVTKSIN--------GDKVRPRIISSFGIGSNYFDTEES 203
W+ G V G V+ ++ Y KY TKSI D+ + + S G+G ++ ES
Sbjct 129 WRMGNVHAGTVTAKSIYYTLKYCTKSITEGPDKDPDDDRKPEKALMSKGLGLSHL--TES 186
Query 204 TLHKLGGQRYQPFMVLNGFQQAMPRYYYNKIFSDVDK 240
+ + F +L G A+PRYY +K+FSD++K
Sbjct 187 MIKYYKDDVSRSFSLLGGTTIALPRYYRDKVFSDIEK 223
>gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=250
Score = 85.9 bits (211), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 66/200 (33%), Positives = 105/200 (53%), Gaps = 35/200 (18%)
Query 67 CLFVTLTFDDDNL--KKFSKD---------TNKAVRLFLDRLRKDYGK-QIRHWFVCEFG 114
LFVTLT+DD+++ +D + + ++LF+ RLRK Y + ++R++ E+G
Sbjct 11 SLFVTLTYDDEHIPTAMIGEDLFKTTVGVVSKRDIQLFMKRLRKKYAQYRLRYFLTSEYG 70
Query 115 TLYGRPHYHGILFDVPQTLIDGYSPDVPGHH--PLLASRWKYGFVFVGYVSDETCSYITK 172
+ GRPHYH ILF P T G H LLA WK GFV ++ + SY+TK
Sbjct 71 SQGGRPHYHMILFGFPFT----------GKHGGDLLAECWKNGFVQAHPLTTKEISYVTK 120
Query 173 YV-TKSINGDKVR------PRIISSF--GIGSNYFDTEESTLHKLGGQRYQPFMVLNGFQ 223
Y+ KS+ D ++ P ++ S GIG ++ + ++L + Y NG +
Sbjct 121 YMYEKSMIPDILKGVKEYQPFMLCSKMPGIGYHFLREQILDFYRLHPRDY--VRAFNGMR 178
Query 224 QAMPRYYYNKIFSDVDKQNI 243
AMPRYY +K++ D K+ +
Sbjct 179 MAMPRYYADKLYDDDMKEYL 198
>gi|495507506|ref|WP_008232152.1| hypothetical protein [Richelia intracellularis]
gi|471331139|emb|CCH66547.1| hypothetical protein RINTHH_3920 [Richelia intracellularis HH01]
Length=306
Score = 85.1 bits (209), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 63/236 (27%), Positives = 109/236 (46%), Gaps = 43/236 (18%)
Query 34 EVPCGYCHSCQKSYNNQYRIRLLYELRKYPPGTCLFVTLTFDD---------DNLKKFSK 84
++PCG+C C + Q+ +R ++E + + FVTLT+++ + +KF K
Sbjct 30 KLPCGHCEGCLLERSRQWAVRCMHEAQLWERNC--FVTLTYEETPPWNSLRHSDFQKFMK 87
Query 85 DTNKAVRLFLDRLRKDYGKQ---IRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDV 141
K + + + GK IR++ E+GT GRPHYH LF+ I+
Sbjct 88 RLRKRFKGHKENIDVRTGKSSYPIRYYMAGEYGTHGGRPHYHACLFNFAFEDIEFLRRTN 147
Query 142 PGHH----PLLASRWKYGFVFVGYVSDETCSYITKYVTKSINGD-------------KVR 184
G + L S W +GF VG V+ E+ +Y+ +YV K +N + +V
Sbjct 148 SGSNLYRSAQLESLWPHGFSSVGDVTFESAAYVARYVMKKMNKEAIEKGQEINWETGEVM 207
Query 185 PRIIS------SFGIGSNYFDTEESTLHKLGGQRYQPFMVLNGFQQAMPRYYYNKI 234
PR+ GIG+N+ D +S + ++++NG + PRYY+ ++
Sbjct 208 PRLPEYNKMSLKPGIGANFIDKYQSDVFP------NDYVIVNGHKAKPPRYYFKRL 257
>gi|575094560|emb|CDL65924.1| unnamed protein product [uncultured bacterium]
Length=320
Score = 84.3 bits (207), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 68/232 (29%), Positives = 106/232 (46%), Gaps = 42/232 (18%)
Query 34 EVPCGYCHSCQ--KSYNNQYRIR---LLYELRKYPPGTCLFVTLTFDDDNLKKFSKDTNK 88
++PCG C C+ +S ++ R LLY+ R Y F+TLT+ ++L F +
Sbjct 44 QIPCGQCIGCRLDRSLDSAVRAHHESLLYD-RNY------FLTLTYSPEHLPPFGSLIPR 96
Query 89 AVRLFLDRLRKDYGKQIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVPGH---- 144
+ LF RLRK G +R+ E+G+ YGRPHYH I+F++P + G
Sbjct 97 DLTLFWKRLRKR-GVSLRYMACGEYGSTYGRPHYHAIIFNLPPLELKQIGTTSTGFPTFI 155
Query 145 HPLLASRWKYGFVFVGYVSDETCSYITKYVTKSINGD-------------------KVRP 185
+++ W GF + VS +TC+Y+ +YVTK I GD K
Sbjct 156 SDVISECWSLGFHTLNPVSFQTCAYVARYVTKKILGDGKQVYEKFDPVTGEVDCRVKEFS 215
Query 186 RIISSFGIGSNYFDTEESTLHKLGGQRYQPFMVLNGFQQAMPRYYYNKIFSD 237
R + GIG +YF +K+ ++N + +PRYY + D
Sbjct 216 RWSTKPGIGHDYFMKYWRDFYKIDC------CLINNKKFKIPRYYDRLLLRD 261
>gi|575094569|emb|CDL65925.1| unnamed protein product [uncultured bacterium]
Length=354
Score = 84.3 bits (207), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 72/241 (30%), Positives = 107/241 (44%), Gaps = 36/241 (15%)
Query 32 YLEVPCGYCHSCQKSYNNQYRIRLLYELRKYPPGTCLFVTLTFDDDNL--------KKFS 83
++E+PCG C SC++ Y + RL+ EL+ + F+TLT+DDD++ + S
Sbjct 67 FIEIPCGKCISCRRRYAALWTDRLMLELQDHKESC--FITLTYDDDHICCVDSPIEENVS 124
Query 84 KDTNKAVRL--FLDRLRK------DYGKQIRHWFVCEFGTLYGRPHYHGILFD-VPQTLI 134
T V L F RLR+ + K+IR++ E+G RPHYH ILF P LI
Sbjct 125 MYTLNKVHLQCFWKRLRQYLVRHVEPEKRIRYFACGEYGDTTFRPHYHAILFGWRPTDLI 184
Query 135 D---GYSPDVPGHHPLLASRWKYGFVFVGYVSDETCSYITKYVTKSING--------DKV 183
+ D LAS W+ G V VG V+ E+C Y+ +Y K G V
Sbjct 185 QFKKNFQNDTLYLSKSLASIWQNGNVMVGDVTPESCRYVARYCLKKATGFDSEIYERLGV 244
Query 184 RPRIISSF---GIGSNYFDTEESTLHKLGGQRYQPFMVLNGFQQAMPRYYYNKIFSDVDK 240
P ++ GI YFD + K G +P Y+ ++ D+D
Sbjct 245 LPEFVTMSRKPGIARKYFDDHYDEIIKYKTINLSTLK--GGMSMQIPPYFI-RLIEDIDS 301
Query 241 Q 241
+
Sbjct 302 E 302
Lambda K H a alpha
0.323 0.141 0.449 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1719536379408