bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-26_CDS_annotation_glimmer3.pl_2_2
Length=243
Score E
Sequences producing significant alignments: (Bits) Value
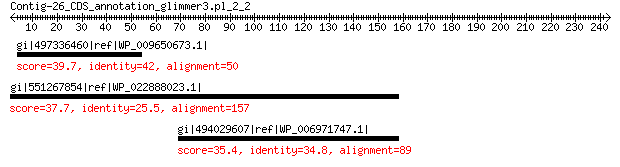
gi|497336460|ref|WP_009650673.1| oxidoreductase 39.7 1.5
gi|551267854|ref|WP_022888023.1| tryptophan synthase subunit beta 37.7 5.3
gi|494029607|ref|WP_006971747.1| hypothetical protein 35.4 8.8
>gi|497336460|ref|WP_009650673.1| oxidoreductase [Campylobacter sp. FOBRC14]
gi|401016189|gb|EJP74957.1| PepSY domain protein [Campylobacter sp. FOBRC14]
Length=388
Score = 39.7 bits (91), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 21/50 (42%), Positives = 28/50 (56%), Gaps = 0/50 (0%)
Query 4 AIGQSHNGEITTLSNRNSHVRALLGSRQRRSEVVVELGGLEGHTAFDNIG 53
AI H+ I L+ RNSHV+ALL SR + SEV+ + +G IG
Sbjct 36 AIISYHDEIIDALNARNSHVKALLPSRMKVSEVLAKFSAQKGEFYLSFIG 85
>gi|551267854|ref|WP_022888023.1| tryptophan synthase subunit beta [Agromyces italicus]
Length=402
Score = 37.7 bits (86), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 40/163 (25%), Positives = 71/163 (44%), Gaps = 22/163 (13%)
Query 1 MVNAIGQSHNGEITTLSNRNSHVRALLGSRQRRSEVVVELGGLEGHTAFDNIGDIVGAVR 60
+ +AI ++ +T ++ N ++ + E+V + + G A + D+ GA+
Sbjct 169 LKDAINEAMRDWVTNVATTN-YLFGTVAGPHPFPEMVRDFQKIIGEEARQQVLDLTGALP 227
Query 61 TCKGSIIHGDVEGVGEVFITEPLIHDEGVGTEGESVNGEVGNTSRDAHVVGNGANRPATR 120
T + + G +G I + DEGVG G GE +T R A + T+
Sbjct 228 TAVAACVGGGSNAIG---IFHAFLDDEGVGLFGFEAGGEGVDTPRHAATI--------TK 276
Query 121 GRSHLGHGILHG------RNPGGLSVLTHELTPMLPYDTVGHE 157
GR G+LHG ++ G ++ +H ++ L Y VG E
Sbjct 277 GRP----GVLHGARSYMLQDDDGQTIESHSISAGLDYPGVGPE 315
>gi|494029607|ref|WP_006971747.1| hypothetical protein [Plesiocystis pacifica]
gi|149819613|gb|EDM79040.1| hypothetical protein PPSIR1_10570 [Plesiocystis pacifica SIR-1]
Length=108
Score = 35.4 bits (80), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 31/89 (35%), Positives = 38/89 (43%), Gaps = 29/89 (33%)
Query 69 GDVEGVGEVFITEPLIHDEGVGTEGESVNGEVGNTSRDAHVVGNGANRPATRGRSHLGHG 128
G E VG++F E L+ D VG ESV E G A RG HLGH
Sbjct 43 GFAEEVGKIF--ESLMVDLVVGVASESVGPEPG----------------AARGLVHLGH- 83
Query 129 ILHGRNPGGLSVLTHELTPMLPYDTVGHE 157
+L HEL P+L DT+G +
Sbjct 84 ----------ELLAHELHPVLAVDTIGRQ 102
Lambda K H a alpha
0.314 0.134 0.386 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1040062966173