bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-26_CDS_annotation_glimmer3.pl_2_5
Length=65
Score E
Sequences producing significant alignments: (Bits) Value
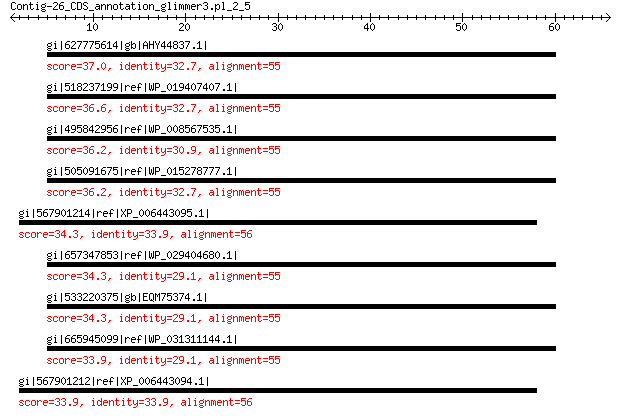
gi|627775614|gb|AHY44837.1| lytic transglycosylase 37.0 0.52
gi|518237199|ref|WP_019407407.1| lytic transglycosylase 36.6 0.81
gi|495842956|ref|WP_008567535.1| lytic transglycosylase 36.2 0.94
gi|505091675|ref|WP_015278777.1| lytic murein transglycosylase 36.2 0.96
gi|567901214|ref|XP_006443095.1| hypothetical protein CICLE_v100... 34.3 3.8
gi|657347853|ref|WP_029404680.1| lytic transglycosylase 34.3 4.1
gi|533220375|gb|EQM75374.1| lytic transglycosylase 34.3 5.0
gi|665945099|ref|WP_031311144.1| lytic transglycosylase 33.9 5.1
gi|567901212|ref|XP_006443094.1| hypothetical protein CICLE_v100... 33.9 6.5
>gi|627775614|gb|AHY44837.1| lytic transglycosylase [Pseudomonas stutzeri]
Length=373
Score = 37.0 bits (84), Expect = 0.52, Method: Composition-based stats.
Identities = 18/55 (33%), Positives = 32/55 (58%), Gaps = 2/55 (4%)
Query 5 QTMEIIDQVYQDMFVETVLKFEVLEKSVDVGSAGRMAHQERLDGMRKLYTKLIKE 59
Q +E++ + ++ MF++ +LK + K+ DV SAG LD MR Y +++ E
Sbjct 26 QQLEMVSEQFEAMFLQQILK--QMRKAGDVLSAGNPMRSRELDTMRDFYDEVLAE 78
>gi|518237199|ref|WP_019407407.1| lytic transglycosylase [Pseudomonas stutzeri]
Length=370
Score = 36.6 bits (83), Expect = 0.81, Method: Composition-based stats.
Identities = 18/55 (33%), Positives = 32/55 (58%), Gaps = 2/55 (4%)
Query 5 QTMEIIDQVYQDMFVETVLKFEVLEKSVDVGSAGRMAHQERLDGMRKLYTKLIKE 59
Q +E++ + ++ MF++ +LK + K+ DV SAG LD MR Y +++ E
Sbjct 26 QQLEMVSEQFEAMFLQQILK--QMRKAGDVLSAGNPMRSRELDTMRDFYDEVLAE 78
>gi|495842956|ref|WP_008567535.1| lytic transglycosylase [Pseudomonas sp. Chol1]
gi|409121014|gb|EKM97201.1| lytic transglycosylase, catalytic [Pseudomonas sp. Chol1]
Length=376
Score = 36.2 bits (82), Expect = 0.94, Method: Composition-based stats.
Identities = 17/55 (31%), Positives = 32/55 (58%), Gaps = 2/55 (4%)
Query 5 QTMEIIDQVYQDMFVETVLKFEVLEKSVDVGSAGRMAHQERLDGMRKLYTKLIKE 59
Q +E++ + ++ MF++ +LK + K+ DV +AG LD MR Y +++ E
Sbjct 26 QQLEVVSEQFEAMFLQQILK--QMRKAGDVLAAGNPMRSRELDTMRDFYDEVLAE 78
>gi|505091675|ref|WP_015278777.1| lytic murein transglycosylase [Pseudomonas stutzeri]
gi|431929211|ref|YP_007242245.1| lytic murein transglycosylase [Pseudomonas stutzeri RCH2]
gi|431827498|gb|AGA88615.1| soluble lytic murein transglycosylase-like protein [Pseudomonas
stutzeri RCH2]
Length=370
Score = 36.2 bits (82), Expect = 0.96, Method: Composition-based stats.
Identities = 18/55 (33%), Positives = 32/55 (58%), Gaps = 2/55 (4%)
Query 5 QTMEIIDQVYQDMFVETVLKFEVLEKSVDVGSAGRMAHQERLDGMRKLYTKLIKE 59
Q +E++ + ++ MF++ +LK + K+ DV SAG LD MR Y +++ E
Sbjct 26 QQLEMVSEQFEAMFLQQILK--QMRKAGDVLSAGNPMRSRELDTMRDFYDEVLAE 78
>gi|567901214|ref|XP_006443095.1| hypothetical protein CICLE_v10024062mg, partial [Citrus clementina]
gi|557545357|gb|ESR56335.1| hypothetical protein CICLE_v10024062mg, partial [Citrus clementina]
Length=303
Score = 34.3 bits (77), Expect = 3.8, Method: Composition-based stats.
Identities = 19/58 (33%), Positives = 33/58 (57%), Gaps = 3/58 (5%)
Query 2 DISQTMEIIDQVYQDMFVETVLKFEVLEK--SVDVGSAGRMAHQERLDGMRKLYTKLI 57
+ +QT+E + + +DM +E +LEK ++++ AG AH LD R+ Y K+I
Sbjct 89 NFNQTLEYVKDI-RDMRMEVCCTLGMLEKQQAIELKKAGLTAHNHNLDTSREYYPKII 145
>gi|657347853|ref|WP_029404680.1| lytic transglycosylase [Pseudomonas stutzeri]
Length=343
Score = 34.3 bits (77), Expect = 4.1, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 31/55 (56%), Gaps = 2/55 (4%)
Query 5 QTMEIIDQVYQDMFVETVLKFEVLEKSVDVGSAGRMAHQERLDGMRKLYTKLIKE 59
Q +E++ + ++ MF++ +LK + K+ DV AG LD MR Y +++ +
Sbjct 26 QQLEVVAEQFEAMFLQQILK--QMRKAGDVLGAGNPMRSRELDTMRDFYDEVLAD 78
>gi|533220375|gb|EQM75374.1| lytic transglycosylase [Pseudomonas stutzeri MF28]
Length=350
Score = 34.3 bits (77), Expect = 5.0, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 31/55 (56%), Gaps = 2/55 (4%)
Query 5 QTMEIIDQVYQDMFVETVLKFEVLEKSVDVGSAGRMAHQERLDGMRKLYTKLIKE 59
Q +E++ + ++ MF++ +LK + K+ DV AG LD MR Y +++ +
Sbjct 34 QQLEVVAEQFEAMFLQQILK--QMRKAGDVLGAGNPMRSRELDTMRDFYDEVLAD 86
>gi|665945099|ref|WP_031311144.1| lytic transglycosylase [Pseudomonas stutzeri]
Length=342
Score = 33.9 bits (76), Expect = 5.1, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 31/55 (56%), Gaps = 2/55 (4%)
Query 5 QTMEIIDQVYQDMFVETVLKFEVLEKSVDVGSAGRMAHQERLDGMRKLYTKLIKE 59
Q +E++ + ++ MF++ +LK + K+ DV AG LD MR Y +++ +
Sbjct 26 QQLEVVAEQFEAMFLQQILK--QMRKAGDVLGAGNPMRSRELDTMRDFYDEVLAD 78
>gi|567901212|ref|XP_006443094.1| hypothetical protein CICLE_v10024495mg [Citrus clementina]
gi|557545356|gb|ESR56334.1| hypothetical protein CICLE_v10024495mg [Citrus clementina]
Length=372
Score = 33.9 bits (76), Expect = 6.5, Method: Composition-based stats.
Identities = 19/58 (33%), Positives = 33/58 (57%), Gaps = 3/58 (5%)
Query 2 DISQTMEIIDQVYQDMFVETVLKFEVLEK--SVDVGSAGRMAHQERLDGMRKLYTKLI 57
+ +QT+E + + +DM +E +LEK ++++ AG AH LD R+ Y K+I
Sbjct 143 NFNQTLEYVKDI-RDMGMEVCCTLGMLEKQQAIELKKAGLTAHNHNLDTSREYYPKII 199
Lambda K H a alpha
0.321 0.136 0.362 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 440996816904