bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-27_CDS_annotation_glimmer3.pl_2_4
Length=238
Score E
Sequences producing significant alignments: (Bits) Value
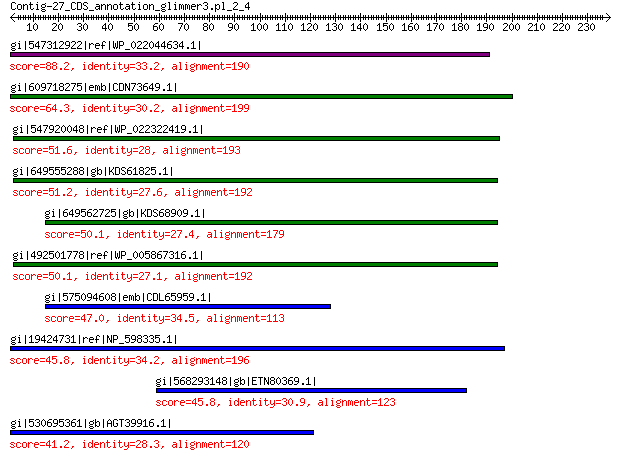
gi|547312922|ref|WP_022044634.1| putative replication initiation... 88.2 3e-17
gi|609718275|emb|CDN73649.1| conserved hypothetical protein 64.3 4e-09
gi|547920048|ref|WP_022322419.1| putative replication protein 51.6 1e-04
gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 51.2 2e-04
gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 50.1 4e-04
gi|492501778|ref|WP_005867316.1| hypothetical protein 50.1 5e-04
gi|575094608|emb|CDL65959.1| unnamed protein product 47.0 0.004
gi|19424731|ref|NP_598335.1| hypothetical protein Sp-4p1 45.8 0.013
gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 45.8 0.013
gi|530695361|gb|AGT39916.1| replication initiator 41.2 0.38
>gi|547312922|ref|WP_022044634.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
gi|524208442|emb|CCZ76638.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
Length=320
Score = 88.2 bits (217), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 63/212 (30%), Positives = 110/212 (52%), Gaps = 41/212 (19%)
Query 1 VRLMEEI-KSNPKNIIFATLTFSEESLKKLEYDEKEPNKAPQKAISLFRKRWWKKYKEPL 59
+RL+ E+ K P +F TLTF+++SL+K D KA+ LF R+ K Y + +
Sbjct 60 IRLLYELRKYPPGTCLFVTLTFNDDSLEKFSKDT-------NKAVRLFLDRFRKVYGKQI 112
Query 60 KHWLITELGHDNTKRIHLHGIIWT--ELTEEQFEKE-----------WGYGWIFFGYEVN 106
+HW + E G + R H HGI++ + + ++ + W YG++F GY V+
Sbjct 113 RHWFVCEFGTLH-GRPHYHGILFNVPQALIDGYDSDMPGHHPLLASCWKYGFVFVGY-VS 170
Query 107 ERTINYIIKYITKR---DEANPEFNGKIFTSKGIGKEYIG--ENSLRR---HRYQDRFTE 158
+ T +YI KY+TK D+ P ++ +S GIG Y+ E+SL + RYQ
Sbjct 171 DETCSYITKYVTKSINGDKVRP----RVISSFGIGSNYLNTEESSLHKLGNQRYQ----- 221
Query 159 ETYRTNSGIKIALPTYYKQKLWTVQEREALRI 190
+ +G + A+P YY K+++ +++ + +
Sbjct 222 -PFMVLNGFQQAMPRYYYNKIFSDVDKQNMVV 252
>gi|609718275|emb|CDN73649.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=265
Score = 64.3 bits (155), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 60/207 (29%), Positives = 100/207 (48%), Gaps = 17/207 (8%)
Query 1 VRLMEEIKSNPKNIIFATLTFSEESLKKLEYDEKEPNKAPQKAISLFRKRWWKKYKEPLK 60
RL EE+K + K+ F TLT+S+ L Y + + LF KR K K +K
Sbjct 41 ARLTEELKVS-KSAHFVTLTYSDV---YLPYSDNGLISLDYRDFQLFMKRARKLQKSKIK 96
Query 61 HWLITELGHDNTKRIHLHGIIWTELTEEQFEKEWGYGWIFFGYEVNERTINYIIKYITKR 120
++L+ E G T R H H I++ + F EW G + G V ++I Y +KY TK
Sbjct 97 YFLVGEYGA-QTYRPHYHAIVFGVENIDAFLGEWRMGNVHAG-TVTAKSIYYTLKYCTKS 154
Query 121 -------DEANPEFNGKIFTSKGIGKEYIGENSLRRHRYQDRFTEETYRTNSGIKIALPT 173
D + K SKG+G ++ E+ ++ Y+D + ++ G IALP
Sbjct 155 ITEGPDKDPDDDRKPEKALMSKGLGLSHLTESMIKY--YKDDVSR-SFSLLGGTTIALPR 211
Query 174 YYKQKLWT-VQEREALRIIKEEKQVKY 199
YY+ K+++ +++ L I + +++Y
Sbjct 212 YYRDKVFSDIEKVHRLVSITDYLEIRY 238
>gi|547920048|ref|WP_022322419.1| putative replication protein [Parabacteroides merdae CAG:48]
gi|524592960|emb|CDD13572.1| putative replication protein [Parabacteroides merdae CAG:48]
Length=278
Score = 51.6 bits (122), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 54/212 (25%), Positives = 105/212 (50%), Gaps = 26/212 (12%)
Query 2 RLMEEIKSNPKNIIFATLTFSEESL--KKLEYDEKEPNKA--PQKAISLFRKRWWKKYKE 57
RL E K P ++ F TLT+ +E L +++ D + N A ++ + LF KR KKY++
Sbjct 29 RLQAEAKEYPLSL-FVTLTYDDEHLPIERIGSDLFQTNVAVVSKRDVQLFMKRLRKKYED 87
Query 58 -PLKHWLITELGHDNTKRIHLHGIIWT-----ELTEEQFEKEWGYGWIFFGYEVNERTIN 111
+++++ +E G N R H H I++ ++ + + W G++ + + + I
Sbjct 88 YKMRYFVTSEYGAKNG-RPHYHMILFGFPFTGKMAGDLLAECWQNGFVQ-AHPLTIKEIA 145
Query 112 YIIKYITKRDEANPE-------FNGKIFTSK--GIGKEYIGENSLRRHRYQDRFTEETYR 162
Y+ KY+ ++ PE + + S+ GIG ++ + + +R R + R
Sbjct 146 YVCKYMYEKSMC-PEILRDEKKYKPFMLCSRNPGIGFGFMKADIIEFYR---RHPRDYVR 201
Query 163 TNSGIKIALPTYYKQKLWTVQEREALRIIKEE 194
+G K+A+P YY KL+ + L+ ++EE
Sbjct 202 AWAGHKMAMPRYYADKLYDDDMKAFLKEMREE 233
>gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649560564|gb|KDS66872.1| hypothetical protein M095_2449 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561011|gb|KDS67298.1| hypothetical protein M095_2409 [Parabacteroides distasonis str.
3999B T(B) 4]
Length=284
Score = 51.2 bits (121), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 53/210 (25%), Positives = 97/210 (46%), Gaps = 24/210 (11%)
Query 2 RLMEEIKSNPKNIIFATLTFSEESLKKLEYDEK----EPNKAPQKAISLFRKRWWKKYKE 57
RL E P ++ F TLT+ +E + E ++ I LF KR KKY +
Sbjct 34 RLQAEADEYPFSL-FVTLTYDDEHIPTAMIGEDLFKTTVGVVSKRDIQLFMKRLRKKYAQ 92
Query 58 -PLKHWLITELGHDNTKRIHLHGIIWT-----ELTEEQFEKEWGYGWIFFGYEVNERTIN 111
L+++L +E G R H H I++ + + + W G++ + + + I+
Sbjct 93 YRLRYFLTSEYG-SQGGRPHYHMILFGFPFTGKHGGDLLAECWKNGFVQ-AHPLTTKEIS 150
Query 112 YIIKYITKRD------EANPEFNGKIFTSK--GIGKEYIGENSLRRHRYQDRFTEETYRT 163
Y+ KY+ ++ + E+ + SK GIG ++ E L +R R + R
Sbjct 151 YVTKYMYEKSMIPDILKGVKEYQPFMLCSKMPGIGYHFLREQILDFYRLHPR---DYVRA 207
Query 164 NSGIKIALPTYYKQKLWTVQEREALRIIKE 193
+G+++A+P YY KL+ +E L+ ++E
Sbjct 208 FNGMRMAMPRYYADKLYDDDMKEYLKELRE 237
>gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=250
Score = 50.1 bits (118), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 49/197 (25%), Positives = 92/197 (47%), Gaps = 23/197 (12%)
Query 15 IFATLTFSEESLKKLEYDEK----EPNKAPQKAISLFRKRWWKKYKE-PLKHWLITELGH 69
+F TLT+ +E + E ++ I LF KR KKY + L+++L +E G
Sbjct 12 LFVTLTYDDEHIPTAMIGEDLFKTTVGVVSKRDIQLFMKRLRKKYAQYRLRYFLTSEYG- 70
Query 70 DNTKRIHLHGIIWT-----ELTEEQFEKEWGYGWIFFGYEVNERTINYIIKYITKRD--- 121
R H H I++ + + + W G++ + + + I+Y+ KY+ ++
Sbjct 71 SQGGRPHYHMILFGFPFTGKHGGDLLAECWKNGFVQ-AHPLTTKEISYVTKYMYEKSMIP 129
Query 122 ---EANPEFNGKIFTSK--GIGKEYIGENSLRRHRYQDRFTEETYRTNSGIKIALPTYYK 176
+ E+ + SK GIG ++ E L +R R + R +G+++A+P YY
Sbjct 130 DILKGVKEYQPFMLCSKMPGIGYHFLREQILDFYRLHPR---DYVRAFNGMRMAMPRYYA 186
Query 177 QKLWTVQEREALRIIKE 193
KL+ +E L+ ++E
Sbjct 187 DKLYDDDMKEYLKELRE 203
>gi|492501778|ref|WP_005867316.1| hypothetical protein [Parabacteroides distasonis]
gi|409230407|gb|EKN23271.1| hypothetical protein HMPREF1059_03256 [Parabacteroides distasonis
CL09T03C24]
Length=284
Score = 50.1 bits (118), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 52/210 (25%), Positives = 95/210 (45%), Gaps = 24/210 (11%)
Query 2 RLMEEIKSNPKNIIFATLTFSEESLKKLEYDE----KEPNKAPQKAISLFRKRWWKKYKE 57
RL E P ++ F TLT+ +E + E ++ I LF KR KKY +
Sbjct 34 RLQAEADEYPFSL-FVTLTYDDEHMPTAMIGEDLFKSTVGVVSKRDIQLFMKRLRKKYDQ 92
Query 58 -PLKHWLITELGHDNTKRIHLHGIIWT-----ELTEEQFEKEWGYGWIFFGYEVNERTIN 111
L+++L +E G R H H I++ + + + W G++ + + + I
Sbjct 93 YRLRYFLTSEYG-SQGGRPHYHMILFGFPFTGKHGGDLLAECWKNGFVQ-AHPLTTKEIA 150
Query 112 YIIKYITKRDEANP------EFNGKIFTSK--GIGKEYIGENSLRRHRYQDRFTEETYRT 163
Y+ KY+ ++ E+ + S+ GIG ++ E L +R R + R
Sbjct 151 YVTKYMYEKSMVPDILKDVKEYQPFMLCSRIPGIGYHFLREQILDFYRLHPR---DYVRA 207
Query 164 NSGIKIALPTYYKQKLWTVQEREALRIIKE 193
+G+++A+P YY KL+ +E L+ ++E
Sbjct 208 FNGMRMAMPRYYADKLYDDDMKEYLKELRE 237
>gi|575094608|emb|CDL65959.1| unnamed protein product [uncultured bacterium]
Length=251
Score = 47.0 bits (110), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 39/130 (30%), Positives = 64/130 (49%), Gaps = 22/130 (17%)
Query 15 IFATLTFSEESLKKLEYDEKEPNKA-PQKAISLFRKRWWKKYK-EPLKHWLITELGHDNT 72
F TLT+++++L YD P + ++ + LF KR K + + ++ +L E G + T
Sbjct 7 CFITLTYNDDNL---PYDVFSPLPSLCKRDVQLFMKRLRKMFSYKQIRFYLCGEYG-EQT 62
Query 73 KRIHLHGIIW-------TEL--TEEQFEKEWGYGWIFFGYEVNERTINYIIKYIT----- 118
R H H II+ T+ + + E W +G + G + N +TI Y+ Y+T
Sbjct 63 HRPHYHAIIFGHDFNADTDFHGSSKTLEHLWQFGNNYVG-QCNPKTIQYVAGYVTKKYVN 121
Query 119 -KRDEANPEF 127
KRD PEF
Sbjct 122 KKRDTITPEF 131
>gi|19424731|ref|NP_598335.1| hypothetical protein Sp-4p1 [Spiroplasma phage 4]
gi|137995|sp|P11334.1|REP_SPV4 RecName: Full=Replication-associated protein ORF2; AltName: Full=Rep
[Spiroplasma phage 4]
Length=320
Score = 45.8 bits (107), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 67/230 (29%), Positives = 100/230 (43%), Gaps = 49/230 (21%)
Query 1 VRLMEEIKSNPKNIIFATLTFSEESLKKLEYDEKEPNKAPQ---KAISLFRKRWWKKYKE 57
VR EIKSNPK+ F TLT+S+E L + PN P+ K I RK + ++
Sbjct 61 VRASLEIKSNPKHNWFVTLTYSDEHL--VYNALGRPNCVPEHITKFIKSLRKYFERRGHI 118
Query 58 PLKHWLITELGHDNTKRIHLH-GIIWTELTEEQFEKE------------------WGYGW 98
+K+ E G TKR+ H I + L + EK W G+
Sbjct 119 GIKYLASNEYG---TKRMRPHYHICFFNLPLDDLEKTIDSQKGYQQWTSKTISRFWDKGF 175
Query 99 IFFGYEVNERTINYIIKYITKR--------DEANPEFNGKIFTSKGIGKEYIGENSLRRH 150
G E+ + NY +Y TK+ + PE K+ SKGIG +Y EN R +
Sbjct 176 HTIG-ELTYHSANYTARYTTKKLGVKDYKALQLVPE---KLRMSKGIGLKYFMENKERIY 231
Query 151 RYQDRFTEETYRTNSGIK-IALPTYYKQKL---WTVQEREALRIIKEEKQ 196
+ T+ GIK +P Y+ +++ W Q+ L IKE+++
Sbjct 232 KEDSVLIS----TDKGIKRFKVPKYFDRRMEREW--QDEFYLDYIKEKRE 275
>gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 [Necator americanus]
Length=345
Score = 45.8 bits (107), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 38/135 (28%), Positives = 60/135 (44%), Gaps = 18/135 (13%)
Query 59 LKHWLITELGHDNTKRIHLHGIIWTELTEEQFEKEWGYGWIFFGYEVNE----RTINYII 114
LK+++ E G R H H II+ + F W G V ++I Y +
Sbjct 93 LKYYMCGEYGSQRF-RPHYHAIIFGVPQDSLFADAWTLNGDSLGGVVVGTVTGKSIAYTM 151
Query 115 KYITK--------RDEANPEFNGKIFTSKGIGKEYIGENSLRRHRYQDRFTEETYRTNSG 166
KYI K RD+ PEF+ SKG+G Y+ + H+ + + G
Sbjct 152 KYIDKSTWKQKHGRDDRVPEFS---LMSKGMGVSYLTPQMVEYHK--EDISRLFCTREGG 206
Query 167 IKIALPTYYKQKLWT 181
+IA+P YY+QK+++
Sbjct 207 SRIAMPRYYRQKIYS 221
>gi|530695361|gb|AGT39916.1| replication initiator [Marine gokushovirus]
Length=289
Score = 41.2 bits (95), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 34/136 (25%), Positives = 54/136 (40%), Gaps = 23/136 (17%)
Query 1 VRLMEEIKSNPKNIIFATLTFSEESLKKLEYDEKEPNKAPQKAISLFRKRWWKKYKEPLK 60
+R + E +++ N F TLTF E + K K P F KR KKY ++
Sbjct 48 IRCVHEAQTHEDNC-FITLTFDNEHIAK----RKNPESLDNTEFQRFMKRLRKKYPHKIR 102
Query 61 HWLITELGHDNTKRIHLHGIIWTE----------------LTEEQFEKEWGYGWIFFGYE 104
+ E G D KR H H +++ ++ + W YG+ G
Sbjct 103 FFHCGEYG-DQNKRPHYHALLFGHDFKDKKLWSNKGDFKLFVSQELAELWPYGFHTIG-A 160
Query 105 VNERTINYIIKYITKR 120
V+ T Y +Y+ K+
Sbjct 161 VSFDTAAYCARYVMKK 176
Lambda K H a alpha
0.316 0.135 0.409 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1002696285300