bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-28_CDS_annotation_glimmer3.pl_2_3
Length=235
Score E
Sequences producing significant alignments: (Bits) Value
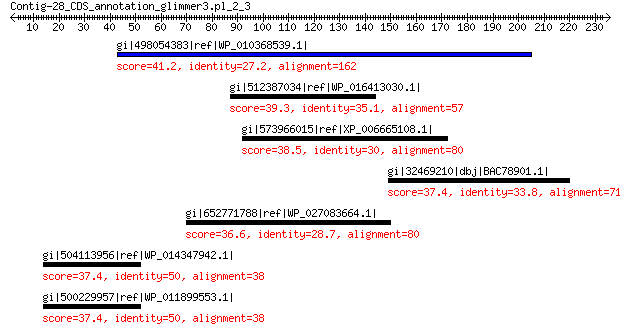
gi|498054383|ref|WP_010368539.1| dTDP-glucose 4,6-dehydratase 41.2 0.34
gi|512387034|ref|WP_016413030.1| putative uncharacterized protein 39.3 1.9
gi|573966015|ref|XP_006665108.1| PREDICTED: uncharacterized prot... 38.5 4.1
gi|32469210|dbj|BAC78901.1| C-type lectin 37.4 4.3
gi|652771788|ref|WP_027083664.1| ribonuclease P 36.6 4.8
gi|504113956|ref|WP_014347942.1| aspartate carbamoyltransferase 37.4 6.5
gi|500229957|ref|WP_011899553.1| aspartate carbamoyltransferase 37.4 7.4
>gi|498054383|ref|WP_010368539.1| dTDP-glucose 4,6-dehydratase [Pseudoalteromonas piscicida]
gi|540272962|gb|ERG32918.1| dTDP-glucose-4,6-dehydratase [Pseudoalteromonas piscicida JCM
20779]
Length=267
Score = 41.2 bits (95), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 44/179 (25%), Positives = 79/179 (44%), Gaps = 27/179 (15%)
Query 43 ITFDAPTCLQ-GRGEIKDEPSLRIS------LINLEIIQMQAEQ-----DHMIMRPKRM- 89
IT+ + T L G+ E +++ +L I L NL ++ AE+ D +I+RP +
Sbjct 82 ITYISSTRLYLGQSETREDSNLVIQGLDQRRLFNLS--KLAAEELCLKSDSLIVRPSNVY 139
Query 90 GTILASSIEPNTIIRERLPKTLHEQLQITLPKGKRRDHFTSLRITITAAGDMIIVIRQGC 149
GT L S + +II++ L K + + + + K +D+ + + D II + +
Sbjct 140 GTALESDLFLPSIIKDALVKKV---VNMYVTKSYSKDY-----VYVGDVADSIIKLIENN 191
Query 150 KPGAIMILQLQKTTVGPFAGILHPAANTLTTW----DLNTFTNYTLCHRSALLDMDGRP 204
G + + + G A IL NT W D + F+ + + LLD + R
Sbjct 192 VKGIVNVASGKNVCAGSIADILEEKTNTKINWHVEEDFDEFSEIDVEKINGLLDYNPRS 250
>gi|512387034|ref|WP_016413030.1| putative uncharacterized protein [Firmicutes bacterium CAG:103]
gi|511628531|emb|CCX45595.1| putative uncharacterized protein [Firmicutes bacterium CAG:103]
Length=697
Score = 39.3 bits (90), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 1/57 (2%)
Query 87 KRMGTILASSIEPNTIIRERLPKTLHEQLQITLPKGKRRDHFTSLRITITAAGDMII 143
+++ I S + + ++RERLPK + Q+Q T+ GKR D + RI A D I
Sbjct 2 EKISDIFGSMVFNDAVMRERLPKETYRQVQATMENGKRLDD-DAARIVANAMKDWAI 57
>gi|573966015|ref|XP_006665108.1| PREDICTED: uncharacterized protein LOC102707274 [Oryza brachyantha]
Length=867
Score = 38.5 bits (88), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 24/83 (29%), Positives = 45/83 (54%), Gaps = 6/83 (7%)
Query 92 ILASSIEPNTIIRERLPKTLHEQLQITLPKG---KRRDHFTSLRITITAAGDMIIVIRQG 148
+L SS+ ++ RE+L + +QL+I LP G + + S R+ AG+ ++ I
Sbjct 527 VLDSSVSISSETREQL---VRKQLEIFLPHGDGVEATGEYDSKRMLTATAGETLVSILSK 583
Query 149 CKPGAIMILQLQKTTVGPFAGIL 171
CKP ++ IL+ + + +G+L
Sbjct 584 CKPISLFILRERNDIIDHLSGML 606
>gi|32469210|dbj|BAC78901.1| C-type lectin [Gymnothorax flavimarginatus]
Length=159
Score = 37.4 bits (85), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 32/74 (43%), Gaps = 10/74 (14%)
Query 149 CKPGAIMILQLQKTTVG--PFA-GILHPAANTLTTWDLNTFTNYTLCHRSALLDMDGRPN 205
C+ + I +LQK G PF G+ + W T YT C+ DG PN
Sbjct 70 CEEDSPFIKELQKAEKGEEPFWIGLTDCHKENIWIWSDGTHVRYTHCN-------DGEPN 122
Query 206 TTHKLHVIHTITGD 219
+ H +HTI GD
Sbjct 123 NLGEEHCVHTIWGD 136
>gi|652771788|ref|WP_027083664.1| ribonuclease P [Lysobacter sp. URHA0019]
Length=135
Score = 36.6 bits (83), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 23/80 (29%), Positives = 38/80 (48%), Gaps = 16/80 (20%)
Query 70 LEIIQMQAEQDHMIMRPKRMGTILASSIEPNTIIRERLPKTLHEQLQITLPKGKRRDHFT 129
L ++ + +D MI R+G ++ ++PN + R R+ + + RD F
Sbjct 30 LPVLALHWNRDDMIA--PRLGLAVSRKVDPNAVGRNRIKRQI-------------RDEFR 74
Query 130 SLRITITAAGDMIIVIRQGC 149
LR T+ A GD +IV R G
Sbjct 75 RLRSTL-APGDYVIVARPGA 93
>gi|504113956|ref|WP_014347942.1| aspartate carbamoyltransferase [Pyrobaculum oguniense]
gi|379005425|ref|YP_005261097.1| aspartate carbamoyltransferase [Pyrobaculum oguniense TE7]
gi|375160878|gb|AFA40490.1| aspartate carbamoyltransferase [Pyrobaculum oguniense TE7]
Length=303
Score = 37.4 bits (85), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 19/38 (50%), Positives = 22/38 (58%), Gaps = 1/38 (3%)
Query 14 LGHIPGSIRTGLPGDLRYNSTANRLLDTIITFDAPTCL 51
GHI G + GL GDLRY T N LL+ + FD T L
Sbjct 147 FGHIDG-LNIGLMGDLRYARTINSLLEALANFDVRTFL 183
>gi|500229957|ref|WP_011899553.1| aspartate carbamoyltransferase [Pyrobaculum arsenaticum]
gi|145590295|ref|YP_001152297.1| aspartate carbamoyltransferase [Pyrobaculum arsenaticum DSM 13514]
gi|172046009|sp|A4WGX8.1|PYRB_PYRAR RecName: Full=Aspartate carbamoyltransferase; AltName: Full=Aspartate
transcarbamylase; Short=ATCase [Pyrobaculum arsenaticum
DSM 13514]
gi|145282063|gb|ABP49645.1| aspartate carbamoyltransferase [Pyrobaculum arsenaticum DSM 13514]
Length=305
Score = 37.4 bits (85), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 19/38 (50%), Positives = 22/38 (58%), Gaps = 1/38 (3%)
Query 14 LGHIPGSIRTGLPGDLRYNSTANRLLDTIITFDAPTCL 51
GHI G + GL GDLRY T N LL+ + FD T L
Sbjct 147 FGHIDG-LNIGLMGDLRYARTINSLLEALANFDVRTFL 183
Lambda K H a alpha
0.323 0.138 0.405 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 971032192080