bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-2_CDS_annotation_glimmer3.pl_2_1
Length=206
Score E
Sequences producing significant alignments: (Bits) Value
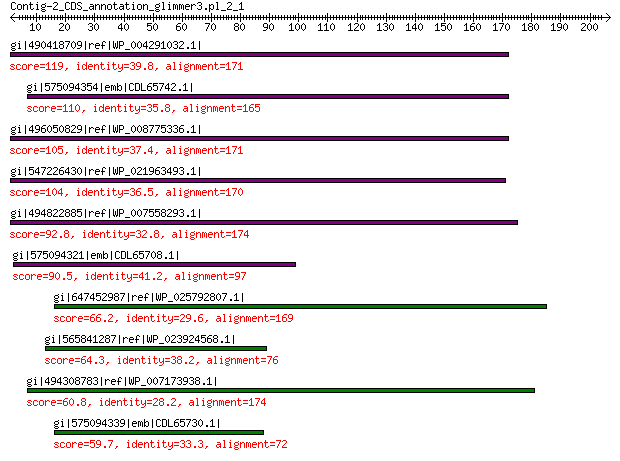
gi|490418709|ref|WP_004291032.1| hypothetical protein 119 1e-27
gi|575094354|emb|CDL65742.1| unnamed protein product 110 1e-24
gi|496050829|ref|WP_008775336.1| hypothetical protein 105 7e-23
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 104 2e-22
gi|494822885|ref|WP_007558293.1| hypothetical protein 92.8 2e-18
gi|575094321|emb|CDL65708.1| unnamed protein product 90.5 2e-17
gi|647452987|ref|WP_025792807.1| hypothetical protein 66.2 2e-09
gi|565841287|ref|WP_023924568.1| hypothetical protein 64.3 7e-09
gi|494308783|ref|WP_007173938.1| hypothetical protein 60.8 9e-08
gi|575094339|emb|CDL65730.1| unnamed protein product 59.7 2e-07
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 119 bits (297), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 68/173 (39%), Positives = 97/173 (56%), Gaps = 11/173 (6%)
Query 1 MAHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRP 60
MA+ LK ++N P R+GFD+ K F+AK GELLPV +PG T+ I+++ FTRT+P
Sbjct 1 MANIMSLKSIRNKPSRNGFDLSFKKNFTAKAGELLPVMVKEVLPGDTFKINLKAFTRTQP 60
Query 61 VQTAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDI--LTALTVSGDLPYC 118
V TAA+ RIREY+DF+ VP DL+W + + QM + P A I +SG++PY
Sbjct 61 VNTAAFARIREYYDFFFVPYDLLWNKANTVLTQMYDN-PQHAVSIDPTRNFVLSGEMPYM 119
Query 119 SLSDLGLSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGN 171
+ + S + ++ KS N FGY R + KL+ L YGN
Sbjct 120 TSEAIASYINALSTASALADYKS--------NYFGYNRSKSSVKLLEYLGYGN 164
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 110 bits (276), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 59/166 (36%), Positives = 93/166 (56%), Gaps = 14/166 (8%)
Query 7 LKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAY 66
+ ++N P R+GFD+ K F+AK GELLPV + +PG +++I+++ FTRT+P+ T+A+
Sbjct 3 MADIKNRPSRNGFDLSFKKNFTAKAGELLPVMTKVVLPGDSFNINLRSFTRTQPLNTSAF 62
Query 67 TRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALT-VSGDLPYCSLSDLGL 125
R+REY+DFY VP + +W FD+ + QM + L T +SG +PY +
Sbjct 63 ARMREYYDFYFVPFEQMWNKFDSCITQMNANVQHASGPTLDDNTPLSGRMPYFT------ 116
Query 126 SCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGN 171
S + + QA A N FG+ R + KL+ L YG+
Sbjct 117 -------SEQIADYLNDQATAARKNPFGFNRSTLTCKLLQYLGYGD 155
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 105 bits (262), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 64/174 (37%), Positives = 100/174 (57%), Gaps = 12/174 (7%)
Query 1 MAHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVY-WDLGIPGCTYDIDIQYFTRTR 59
MA+ LK L+N R+GFD+ +K F+AK GELLPV W++ +PG + ID++ FTRT+
Sbjct 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKPGELLPVKCWEV-LPGDKWSIDLKSFTRTQ 59
Query 60 PVQTAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTAL--TVSGDLPY 117
P+ TAA+ R+REY+DFY VP +L+W + + QM + P A + + ++G +P
Sbjct 60 PLNTAAFARMREYYDFYFVPYNLLWNKANTVLTQMYDN-PQHATSYIPSANQALAGVMPN 118
Query 118 CSLSDLGLSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGN 171
+ G++ + + V + S++ N FGY R KL+ L YGN
Sbjct 119 VTCK--GIADYLNLVAPDVTTTNSYE-----KNYFGYSRSLGTAKLLEYLGYGN 165
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 104 bits (260), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 62/170 (36%), Positives = 97/170 (57%), Gaps = 9/170 (5%)
Query 1 MAHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRP 60
M+ L L+N R+GFD+ KN F+AK GELLP+ PG ++I Q FTRT+P
Sbjct 1 MSSVMSLTALKNSVKRNGFDLSFKNAFTAKVGELLPIMCKEVYPGDKFNIRGQAFTRTQP 60
Query 61 VQTAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSL 120
V +AAY+R+REY+DFY VP L+W M + P A D+++++ +S P+ +
Sbjct 61 VNSAAYSRLREYYDFYFVPYRLLWNMAPTFFTNMPD--PHHAADLVSSVNLSQRHPWFTF 118
Query 121 SDLGLSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYG 170
D+ + S+S + + +Q N FG+ R +++ KL++ LNYG
Sbjct 119 FDI-MEYLGNLNSLS-GAYEKYQ-----KNFFGFSRVELSVKLLNYLNYG 161
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 92.8 bits (229), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 57/179 (32%), Positives = 91/179 (51%), Gaps = 24/179 (13%)
Query 1 MAHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRP 60
MA+ +K ++N P R+G+D+ K F+AK G L+PV+W +P + ++ F RT+P
Sbjct 8 MANIMSMKSVRNKPTRAGYDLTQKINFTAKAGSLIPVWWTPVLPFDDLNATVKSFVRTQP 67
Query 61 VQTAAYTRIREYFDFYAVPIDLIWKSFDASVIQM-----GETAPVQAKDILTALTVSGDL 115
+ TAA+ R+R YFDFY VP +W F ++ QM + PV A ++ +S +L
Sbjct 68 LNTAAFARMRGYFDFYFVPFRQMWNKFPTAITQMRTNLLHASGPVLADNV----PLSDEL 123
Query 116 PYCSLSDLGLSCFFASGSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIP 174
PY + + A +S+ K N FGY R + ++ L YG+ P
Sbjct 124 PYFTAEQV------ADYIVSLADSK---------NQFGYYRAWLVCIILEYLGYGDFYP 167
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 90.5 bits (223), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 40/97 (41%), Positives = 61/97 (63%), Gaps = 0/97 (0%)
Query 2 AHFTGLKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPV 61
++ GL L+N P R+ FD+ +N+F+AK GELLP + PG + + YFTRT P+
Sbjct 5 SNIMGLHGLKNKPSRNSFDLSHRNMFTAKVGELLPCFVQELNPGDSVKVSSSYFTRTAPL 64
Query 62 QTAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETA 98
Q+ A+TR+RE ++ VP +WK FD+ V+ M + A
Sbjct 65 QSNAFTRLRENVQYFFVPYSALWKYFDSQVLNMTKNA 101
>gi|647452987|ref|WP_025792807.1| hypothetical protein [Prevotella histicola]
Length=584
Score = 66.2 bits (160), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 50/176 (28%), Positives = 85/176 (48%), Gaps = 29/176 (16%)
Query 16 RSGFDIGAKNVFSAKCGELLPV-YWDLGIPGCTYDIDIQYFTRTRPVQTAAYTRIREYFD 74
R+GFD+ ++ +FSAK G+LLP+ W++ P + +Q RT + TA+Y R++EY+
Sbjct 10 RNGFDLSSRRIFSAKAGQLLPIGCWEVN-PSEHFKFSVQDLVRTTTLNTASYARMKEYYH 68
Query 75 FYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSLSDLGLSCFFASGSM 134
F+ V +W+ FD ++ G P A L + +G Y +
Sbjct 69 FFFVSYRSLWQWFDQFIV--GTNNPHSA---LNGVKKNGTTNYNQICS------------ 111
Query 135 SVPS------LKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNNMPALNIGN 184
SVP+ + + ++ + F Y G KL++MLNYG + N +N+ N
Sbjct 112 SVPTFDLGKLITRLKTSDMDSQGFNYSEGAA--KLLNMLNYG--VTNKGKFMNLEN 163
>gi|565841287|ref|WP_023924568.1| hypothetical protein [Prevotella nigrescens]
gi|564729907|gb|ETD29851.1| hypothetical protein HMPREF1173_00033 [Prevotella nigrescens
CC14M]
Length=656
Score = 64.3 bits (155), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 29/78 (37%), Positives = 47/78 (60%), Gaps = 2/78 (3%)
Query 13 HPH--RSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAYTRIR 70
HP+ R+G+D+ ++ +FSA G LLP+ PG + I +Q R +P+ TAA+ R +
Sbjct 10 HPNLNRNGYDLSSRRIFSAPAGALLPIATWEANPGEKFRISVQDLVRAQPLNTAAFARCK 69
Query 71 EYFDFYAVPIDLIWKSFD 88
EY+ F+ VP +W+ D
Sbjct 70 EYYHFFFVPYKSLWQHSD 87
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 60.8 bits (146), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 49/178 (28%), Positives = 83/178 (47%), Gaps = 13/178 (7%)
Query 7 LKQLQNHPHRSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAY 66
+K + + +R+ FD+ +++F+A G LLPV IP +I+ Q F RT P+ TAA+
Sbjct 8 IKATRPNRNRNAFDLSQRHLFTAHAGMLLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAF 67
Query 67 TRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSLSDL--G 124
+R ++F+ VP +W FD + M + K I T +PY ++ +
Sbjct 68 ASMRGVYEFFFVPYHQLWAQFDQFITGMNDFHSSANKSIQGG-TSPLQVPYFNVDSVFNS 126
Query 125 LSCFFASGSMSVPSLKSWQANNAYA--NIFGYIRGDVNYKLIHMLNYGNIIPNNMPAL 180
L+ SGS S L+ A+ ++ GY R ++G P+N+ L
Sbjct 127 LNTGKESGSGSTDDLQYKFKYGAFRLLDLLGYGR--------KFDSFGTAYPDNVSGL 176
>gi|575094339|emb|CDL65730.1| unnamed protein product [uncultured bacterium]
Length=588
Score = 59.7 bits (143), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 46/72 (64%), Gaps = 0/72 (0%)
Query 16 RSGFDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAYTRIREYFDF 75
++GFD+ ++ F++ G+LLPV++D PG I FTRT+P+++ A R+ E+ ++
Sbjct 16 KNGFDMSQRHPFTSSVGQLLPVFYDYLNPGDKIRISANLFTRTQPMKSTAMARLTEHIEY 75
Query 76 YAVPIDLIWKSF 87
+ VP + ++ F
Sbjct 76 FFVPFEQMFSLF 87
Lambda K H a alpha
0.321 0.137 0.436 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 692426810790