bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-2_CDS_annotation_glimmer3.pl_2_5
Length=71
Score E
Sequences producing significant alignments: (Bits) Value
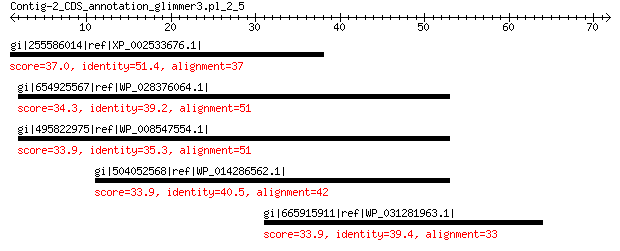
gi|255586014|ref|XP_002533676.1| pentatricopeptide repeat-contai... 37.0 0.76
gi|654925567|ref|WP_028376064.1| mechanosensitive ion channel pr... 34.3 4.5
gi|495822975|ref|WP_008547554.1| glutamine synthetase 33.9 7.5
gi|504052568|ref|WP_014286562.1| glutamine synthetase 33.9 7.9
gi|665915911|ref|WP_031281963.1| hypothetical protein 33.9 8.0
>gi|255586014|ref|XP_002533676.1| pentatricopeptide repeat-containing protein, putative [Ricinus
communis]
gi|223526427|gb|EEF28706.1| pentatricopeptide repeat-containing protein, putative [Ricinus
communis]
Length=726
Score = 37.0 bits (84), Expect = 0.76, Method: Composition-based stats.
Identities = 19/44 (43%), Positives = 29/44 (66%), Gaps = 7/44 (16%)
Query 1 MKENFYFVL-----YLGTSVLNVY--AGSQSNARAYFRRWIEKN 37
+ NF+++L +LGTS+LN+Y GS S+AR YF R + K+
Sbjct 455 LTRNFFYILEGDNIHLGTSILNMYIRCGSISSAREYFNRMVAKD 498
>gi|654925567|ref|WP_028376064.1| mechanosensitive ion channel protein MscS [Leeuwenhoekiella sp.
Hel_I_48]
Length=359
Score = 34.3 bits (77), Expect = 4.5, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 31/52 (60%), Gaps = 3/52 (6%)
Query 2 KENFYFVLYLG-TSVLNVYAGSQSNARAYFRRWIEKNLPVYYHESDFWFLRY 52
+ENFY VLYLG +VL ++A S + +F+R I+ + + + FLRY
Sbjct 85 RENFYIVLYLGIVAVLTLFAASL--VQTWFKRSIKHRVANNEDTTSYKFLRY 134
>gi|495822975|ref|WP_008547554.1| glutamine synthetase [Pseudovibrio sp. JE062]
gi|211960451|gb|EEA95647.1| glutamine synthetase, putative [Pseudovibrio sp. JE062]
Length=468
Score = 33.9 bits (76), Expect = 7.5, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 28/51 (55%), Gaps = 6/51 (12%)
Query 2 KENFYFVLYLGTSVLNVYAGSQSNARAYFRRWIEKNLPVYYHESDFWFLRY 52
+E+ + LG ++ + YA S+ N FR WIEK++ + F FLRY
Sbjct 420 RESEFVQQALGVALADQYARSRENELKAFRAWIEKDI------TSFEFLRY 464
>gi|504052568|ref|WP_014286562.1| glutamine synthetase [Pseudovibrio sp. FO-BEG1]
gi|374332548|ref|YP_005082732.1| glutamine synthetase [Pseudovibrio sp. FO-BEG1]
gi|359345336|gb|AEV38710.1| glutamine synthetase, catalytic region [Pseudovibrio sp. FO-BEG1]
Length=468
Score = 33.9 bits (76), Expect = 7.9, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 24/42 (57%), Gaps = 6/42 (14%)
Query 11 LGTSVLNVYAGSQSNARAYFRRWIEKNLPVYYHESDFWFLRY 52
LG ++ + YA S+ N FR WIEK++ + F FLRY
Sbjct 429 LGVALADQYARSRENELKAFRAWIEKDI------TSFEFLRY 464
>gi|665915911|ref|WP_031281963.1| hypothetical protein, partial [Corynebacterium-like bacterium
B27]
Length=483
Score = 33.9 bits (76), Expect = 8.0, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 19/33 (58%), Gaps = 0/33 (0%)
Query 31 RRWIEKNLPVYYHESDFWFLRYECFSDFLASRY 63
RR+I P +H D+W+L Y FS+ +RY
Sbjct 193 RRYIAHECPEVFHWGDWWYLVYSEFSEGFTTRY 225
Lambda K H a alpha
0.329 0.142 0.465 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 432762933120