bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-2_CDS_annotation_glimmer3.pl_2_6
Length=365
Score E
Sequences producing significant alignments: (Bits) Value
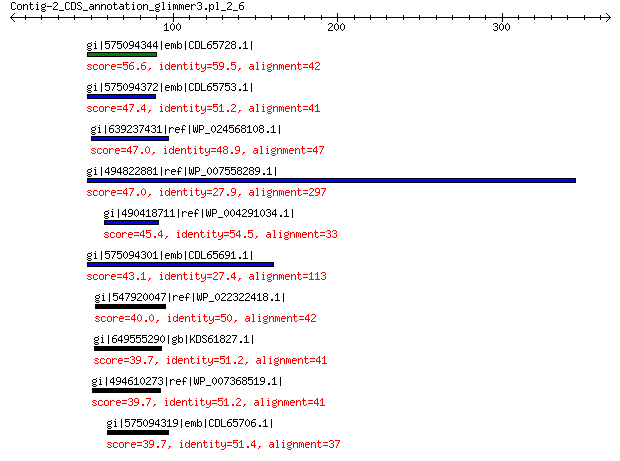
gi|575094344|emb|CDL65728.1| unnamed protein product 56.6 1e-05
gi|575094372|emb|CDL65753.1| unnamed protein product 47.4 0.009
gi|639237431|ref|WP_024568108.1| hypothetical protein 47.0 0.010
gi|494822881|ref|WP_007558289.1| hypothetical protein 47.0 0.013
gi|490418711|ref|WP_004291034.1| hypothetical protein 45.4 0.042
gi|575094301|emb|CDL65691.1| unnamed protein product 43.1 0.28
gi|547920047|ref|WP_022322418.1| putative uncharacterized protein 40.0 1.5
gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 39.7 2.3
gi|494610273|ref|WP_007368519.1| hypothetical protein 39.7 2.7
gi|575094319|emb|CDL65706.1| unnamed protein product 39.7 3.3
>gi|575094344|emb|CDL65728.1| unnamed protein product [uncultured bacterium]
Length=368
Score = 56.6 bits (135), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/42 (60%), Positives = 31/42 (74%), Gaps = 0/42 (0%)
Query 48 DFNAREAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPY 89
++ ++EAQK RDFQL+MWNR NEYN P Q K EAG NP+
Sbjct 51 EWQSQEAQKQRDFQLDMWNRNNEYNKPDEQMKRLEEAGINPW 92
>gi|575094372|emb|CDL65753.1| unnamed protein product [uncultured bacterium]
Length=385
Score = 47.4 bits (111), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 21/41 (51%), Positives = 29/41 (71%), Gaps = 0/41 (0%)
Query 48 DFNAREAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNP 88
+FNA EA+K R FQ ++ R +NSP+NQ K+ A+AG NP
Sbjct 72 EFNASEAEKNRAFQKSLYERSLSWNSPSNQLKMMADAGLNP 112
>gi|639237431|ref|WP_024568108.1| hypothetical protein [Elizabethkingia anophelis]
Length=287
Score = 47.0 bits (110), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 23/47 (49%), Positives = 29/47 (62%), Gaps = 0/47 (0%)
Query 50 NAREAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYLGYDSNT 96
N + A++ R F L+MWNR NEYN+P Q + EAG NP L Y T
Sbjct 23 NKKIARENRAFALDMWNRNNEYNTPLAQMQRLKEAGLNPNLMYGQGT 69
>gi|494822881|ref|WP_007558289.1| hypothetical protein [Bacteroides plebeius]
gi|198272097|gb|EDY96366.1| hypothetical protein BACPLE_00802 [Bacteroides plebeius DSM 17135]
Length=344
Score = 47.0 bits (110), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 83/311 (27%), Positives = 132/311 (42%), Gaps = 65/311 (21%)
Query 48 DFNAREAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYLGYDSNT-GVagstgsts 106
++N + ++ + + +MWN +NEYNS ++QRK EAG NPY+ D + G A S S +
Sbjct 56 EYNREQLERQIEQEWDMWNAENEYNSASSQRKRLEEAGLNPYMMMDGGSAGSASSMTSPA 115
Query 107 qaqaasP----LSLNPEVYSELGSQLGRAGQMIYQERESNARTKA---LQGDADVARAQA 159
A P ++ P S L G A + I A KA ++G + Q
Sbjct 116 AQPAVVPQMQGATMQPADMSGLSGLRGIASEFI-------ATLKAQEDIRGQQLINEGQE 168
Query 160 LQVFSNVDWGKLSPDYKKWMRETGLQRAQLDYDTNKQNLQNLRWSNLIQMAERTNLLLSA 219
++ D KL D +K E+G R+Q T Q++ N R+ R +L S
Sbjct 169 IENQYKAD--KLLADLEKTRTESGFVRSQ----TKGQDIMN-RF--------RPEMLSSE 213
Query 220 NTKR---TLNKYLDQSEQTRINVMAAQYYDLMAAGHLKYQQCKESIAKQILYSKQGSWYD 276
+R T+ L Q N+ A Q+Y ++ QQ K++I +Q
Sbjct 214 IRQRKTDTMFTQLRAHGQMLANLSAYQWYKVLP------QQIKQTINEQ----------- 256
Query 277 SMANKNNLDYRNALALADDYIAAMSTQYESQTAYNMG---FGQKAQEAGRRDAESKSFKS 333
M NN+ + L TQ + T N F + A+E + D ES S+K
Sbjct 257 -MVRINNMKLQGNL-----------TQAQINTEINKAVTEFMKGAREQQQFDFESDSYKD 304
Query 334 LIDRWNYNKRY 344
+D+ + R+
Sbjct 305 RLDQIKADLRH 315
>gi|490418711|ref|WP_004291034.1| hypothetical protein [Bacteroides eggerthii]
gi|217986638|gb|EEC52972.1| hypothetical protein BACEGG_02723 [Bacteroides eggerthii DSM
20697]
Length=368
Score = 45.4 bits (106), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 18/33 (55%), Positives = 23/33 (70%), Gaps = 0/33 (0%)
Query 58 RDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYL 90
+ +Q EMWN+QNEYN P+ QR AG NPY+
Sbjct 86 KAYQTEMWNKQNEYNDPSAQRARLEAAGLNPYM 118
>gi|575094301|emb|CDL65691.1| unnamed protein product [uncultured bacterium]
Length=437
Score = 43.1 bits (100), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 31/113 (27%), Positives = 51/113 (45%), Gaps = 15/113 (13%)
Query 48 DFNAREAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYLGYDSNTGVagstgstsq 107
+F +E RDF +MW N+YN+P Q++ +AG NPY+ D N G A
Sbjct 53 NFTHQENALQRDFARQMWKDTNDYNTPIAQKQRLEQAGMNPYVNMD-NAGTAQ------- 104
Query 108 aqaasPLSLNPEVYSELGSQLGRAGQMIYQERESNARTKALQGDADVARAQAL 160
S+ P V G+ G G + + A ++A + A+ R++ +
Sbjct 105 -------SVTPAVGGSAGATTGTQGAAQIAQGNAIASSQAAKNLAEAERSREM 150
>gi|547920047|ref|WP_022322418.1| putative uncharacterized protein [Parabacteroides merdae CAG:48]
gi|524592959|emb|CDD13571.1| putative uncharacterized protein [Parabacteroides merdae CAG:48]
Length=259
Score = 40.0 bits (92), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 21/42 (50%), Positives = 25/42 (60%), Gaps = 0/42 (0%)
Query 53 EAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYLGYDS 94
E +KA +EMWN QN+YNSP Q +AG NP L Y S
Sbjct 14 ENEKAYARSVEMWNMQNQYNSPTAQMSRLRQAGLNPNLVYGS 55
>gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649557306|gb|KDS63785.1| hypothetical protein M095_3404 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649559158|gb|KDS65545.1| hypothetical protein M096_4689 [Parabacteroides distasonis str.
3999B T(B) 6]
gi|649560567|gb|KDS66875.1| hypothetical protein M095_2448 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561016|gb|KDS67303.1| hypothetical protein M095_2410 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649562727|gb|KDS68911.1| hypothetical protein M096_3341 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=288
Score = 39.7 bits (91), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 21/41 (51%), Positives = 24/41 (59%), Gaps = 0/41 (0%)
Query 52 REAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYLGY 92
+E +KA L+MWN QNEYNSP Q AG NP L Y
Sbjct 48 QENEKAYQRSLKMWNLQNEYNSPTQQMARIRAAGLNPNLVY 88
>gi|494610273|ref|WP_007368519.1| hypothetical protein [Prevotella multiformis]
gi|324988545|gb|EGC20508.1| hypothetical protein HMPREF9141_0987 [Prevotella multiformis
DSM 16608]
Length=437
Score = 39.7 bits (91), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 21/49 (43%), Positives = 27/49 (55%), Gaps = 8/49 (16%)
Query 51 AREA--------QKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYLG 91
AREA Q+ F MWN+ N+YNSPA Q + +AG NPY+
Sbjct 104 AREANQNQYQMFQEQNAFNERMWNQMNQYNSPAAQMQRYTDAGINPYIA 152
>gi|575094319|emb|CDL65706.1| unnamed protein product [uncultured bacterium]
Length=396
Score = 39.7 bits (91), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 19/37 (51%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 60 FQLEMWNRQNEYNSPANQRKLRAEAGYNPYLGYDSNT 96
F MWN QNEYN P QR AG NPYL D +
Sbjct 73 FNERMWNLQNEYNRPDMQRARLEAAGLNPYLMMDGGS 109
Lambda K H a alpha
0.313 0.127 0.365 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2216296179792