bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_3
Length=365
Score E
Sequences producing significant alignments: (Bits) Value
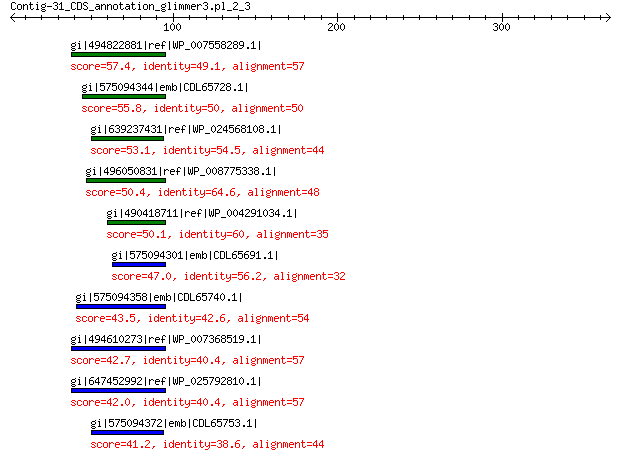
gi|494822881|ref|WP_007558289.1| hypothetical protein 57.4 6e-06
gi|575094344|emb|CDL65728.1| unnamed protein product 55.8 2e-05
gi|639237431|ref|WP_024568108.1| hypothetical protein 53.1 1e-04
gi|496050831|ref|WP_008775338.1| predicted protein 50.4 0.001
gi|490418711|ref|WP_004291034.1| hypothetical protein 50.1 0.001
gi|575094301|emb|CDL65691.1| unnamed protein product 47.0 0.015
gi|575094358|emb|CDL65740.1| unnamed protein product 43.5 0.17
gi|494610273|ref|WP_007368519.1| hypothetical protein 42.7 0.29
gi|647452992|ref|WP_025792810.1| hypothetical protein 42.0 0.62
gi|575094372|emb|CDL65753.1| unnamed protein product 41.2 0.88
>gi|494822881|ref|WP_007558289.1| hypothetical protein [Bacteroides plebeius]
gi|198272097|gb|EDY96366.1| hypothetical protein BACPLE_00802 [Bacteroides plebeius DSM 17135]
Length=344
Score = 57.4 bits (137), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 28/57 (49%), Positives = 38/57 (67%), Gaps = 0/57 (0%)
Query 38 KSANAMNYKINQMNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPY 94
+S N N +I QM+NE+N + ++ + DMWN NEYNS S+ R+RL EAG NPY
Sbjct 41 ESTNQANIQIAQMSNEYNREQLERQIEQEWDMWNAENEYNSASSQRKRLEEAGLNPY 97
>gi|575094344|emb|CDL65728.1| unnamed protein product [uncultured bacterium]
Length=368
Score = 55.8 bits (133), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/50 (50%), Positives = 33/50 (66%), Gaps = 2/50 (4%)
Query 45 YKINQMNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPY 94
+ + QM E+ + EAQK RD+ DMWN+ NEYN P +RL EAG NP+
Sbjct 45 FALGQM--EWQSQEAQKQRDFQLDMWNRNNEYNKPDEQMKRLEEAGINPW 92
>gi|639237431|ref|WP_024568108.1| hypothetical protein [Elizabethkingia anophelis]
Length=287
Score = 53.1 bits (126), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/44 (55%), Positives = 30/44 (68%), Gaps = 0/44 (0%)
Query 50 MNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNP 93
MNN N A+++R + DMWN+ NEYN+P A QRL EAG NP
Sbjct 18 MNNSSNKKIARENRAFALDMWNRNNEYNTPLAQMQRLKEAGLNP 61
>gi|496050831|ref|WP_008775338.1| predicted protein [Bacteroides sp. 2_2_4]
gi|229448895|gb|EEO54686.1| hypothetical protein BSCG_01611 [Bacteroides sp. 2_2_4]
Length=381
Score = 50.4 bits (119), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 31/70 (44%), Positives = 33/70 (47%), Gaps = 22/70 (31%)
Query 47 INQMNNEFNAAEAQKSRDYMT----------------------DMWNKTNEYNSPSAMRQ 84
I QMNNEFN QK DY T DM+N TNEYNS SA R+
Sbjct 41 IAQMNNEFNERMLQKQMDYNTLAYDQQVSDQWSFYNDAKQNAWDMFNATNEYNSASAQRE 100
Query 85 RLVEAGYNPY 94
R AG NPY
Sbjct 101 RYEAAGLNPY 110
>gi|490418711|ref|WP_004291034.1| hypothetical protein [Bacteroides eggerthii]
gi|217986638|gb|EEC52972.1| hypothetical protein BACEGG_02723 [Bacteroides eggerthii DSM
20697]
Length=368
Score = 50.1 bits (118), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/35 (60%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 60 QKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPY 94
+ ++ Y T+MWNK NEYN PSA R RL AG NPY
Sbjct 83 EDNKAYQTEMWNKQNEYNDPSAQRARLEAAGLNPY 117
>gi|575094301|emb|CDL65691.1| unnamed protein product [uncultured bacterium]
Length=437
Score = 47.0 bits (110), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 18/32 (56%), Positives = 23/32 (72%), Gaps = 0/32 (0%)
Query 63 RDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPY 94
RD+ MW TN+YN+P A +QRL +AG NPY
Sbjct 63 RDFARQMWKDTNDYNTPIAQKQRLEQAGMNPY 94
>gi|575094358|emb|CDL65740.1| unnamed protein product [uncultured bacterium]
Length=328
Score = 43.5 bits (101), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 23/54 (43%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 41 NAMNYKINQMNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPY 94
N+ N +I QMNNE++ +K Y T+MW K +YNS Q+ +AG NPY
Sbjct 27 NSTNMQIAQMNNEWSERMMEKQMAYNTEMWEKVADYNSLPNKMQQARDAGVNPY 80
>gi|494610273|ref|WP_007368519.1| hypothetical protein [Prevotella multiformis]
gi|324988545|gb|EGC20508.1| hypothetical protein HMPREF9141_0987 [Prevotella multiformis
DSM 16608]
Length=437
Score = 42.7 bits (99), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 30/57 (53%), Gaps = 11/57 (19%)
Query 38 KSANAMNYKINQMNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPY 94
+ AN Y++ Q N FN MWN+ N+YNSP+A QR +AG NPY
Sbjct 105 REANQNQYQMFQEQNAFN-----------ERMWNQMNQYNSPAAQMQRYTDAGINPY 150
>gi|647452992|ref|WP_025792810.1| hypothetical protein [Prevotella histicola]
Length=424
Score = 42.0 bits (97), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 31/57 (54%), Gaps = 11/57 (19%)
Query 38 KSANAMNYKINQMNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPY 94
+ N MNY++ Q +N FN M+++ N YN+PSA QR EAG NPY
Sbjct 38 RETNQMNYQLFQESNAFNE-----------KMYHEANAYNTPSAQMQRYAEAGINPY 83
>gi|575094372|emb|CDL65753.1| unnamed protein product [uncultured bacterium]
Length=385
Score = 41.2 bits (95), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 17/44 (39%), Positives = 30/44 (68%), Gaps = 0/44 (0%)
Query 50 MNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNP 93
+ NEFNA+EA+K+R + ++ ++ +NSPS + + +AG NP
Sbjct 69 LQNEFNASEAEKNRAFQKSLYERSLSWNSPSNQLKMMADAGLNP 112
Lambda K H a alpha
0.311 0.121 0.348 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2216296179792