bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_5
Length=166
Score E
Sequences producing significant alignments: (Bits) Value
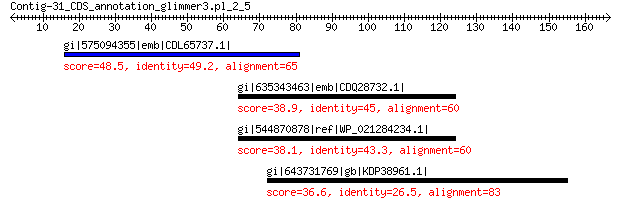
gi|575094355|emb|CDL65737.1| unnamed protein product 48.5 6e-04
gi|635343463|emb|CDQ28732.1| Dipeptide-binding protein DppE prec... 38.9 1.1
gi|544870878|ref|WP_021284234.1| hypothetical protein 38.1 2.2
gi|643731769|gb|KDP38961.1| hypothetical protein JCGZ_00718 36.6 6.9
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 48.5 bits (114), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 32/65 (49%), Positives = 41/65 (63%), Gaps = 0/65 (0%)
Query 16 CLRPRRVTNPYTADVLFSPcgccaacvankanvataYVQNMASYFKFCYFVTLTYADTFL 75
CL P+RV NPY D L PCG C AC +KA+ +Q AS KFC F TLTYA+T++
Sbjct 11 CLEPKRVFNPYLNDWLLVPCGKCRACQCSKASRYKLQIQLEASQHKFCIFGTLTYANTYI 70
Query 76 PMVDV 80
P + +
Sbjct 71 PRLSL 75
>gi|635343463|emb|CDQ28732.1| Dipeptide-binding protein DppE precursor [Halobacillus trueperi]
gi|635346794|emb|CDQ21028.1| Dipeptide-binding protein DppE precursor [Halobacillus dabanensis]
gi|662981198|emb|CDQ24908.1| Dipeptide-binding protein DppE precursor [Halobacillus karajensis]
Length=563
Score = 38.9 bits (89), Expect = 1.1, Method: Composition-based stats.
Identities = 27/60 (45%), Positives = 35/60 (58%), Gaps = 3/60 (5%)
Query 64 YFVTLTYADTFLPMVDVCAVERTGNRYLEYADTLLPSSDPRDLKEEWYHDRDLSLDSNEN 123
YF +LT TFLPM + VE G++Y ADTLL S+ P L EW H +L+ NE+
Sbjct 196 YFESLTTFGTFLPM-NQAFVEEHGDQYALEADTLL-SNGPFVLT-EWNHGEGWTLEKNED 252
>gi|544870878|ref|WP_021284234.1| hypothetical protein [Clostridium sp. BL8]
gi|530689129|gb|EQB87274.1| hypothetical protein M918_09890 [Clostridium sp. BL8]
Length=564
Score = 38.1 bits (87), Expect = 2.2, Method: Composition-based stats.
Identities = 26/60 (43%), Positives = 31/60 (52%), Gaps = 3/60 (5%)
Query 64 YFVTLTYADTFLPMVDVCAVERTGNRYLEYADTLLPSSDPRDLKEEWYHDRDLSLDSNEN 123
YF+ LTY F+P VE+ Y A TLL S P LKE W HD + L+ NEN
Sbjct 183 YFMNLTYFTLFMPQRQD-MVEKNAKNYGADAGTLL-SCGPFKLKE-WVHDSKIVLEKNEN 239
>gi|643731769|gb|KDP38961.1| hypothetical protein JCGZ_00718 [Jatropha curcas]
Length=806
Score = 36.6 bits (83), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 22/84 (26%), Positives = 41/84 (49%), Gaps = 7/84 (8%)
Query 72 DTFLPMVDVCAVERTGNRYLEYADTLLPSSDPRDLKEEWYHDRDLSLDSNENLVRQQYEI 131
D ++ + D+ + +R L+ + LLP DP D E L +NE++V Q +
Sbjct 576 DVYIDVPDIIDISHMRSRGLQPGEQLLPEGDPSDEVES------KQLVANEDIVSQLVSM 629
Query 132 GFKYVP-RKVSVKVKNSTVYRSFD 154
GF Y+ +K ++ N+ V + +
Sbjct 630 GFNYLHCQKAAINTLNTGVEEAMN 653
Lambda K H a alpha
0.323 0.139 0.426 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 429014905530