bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_6
Length=236
Score E
Sequences producing significant alignments: (Bits) Value
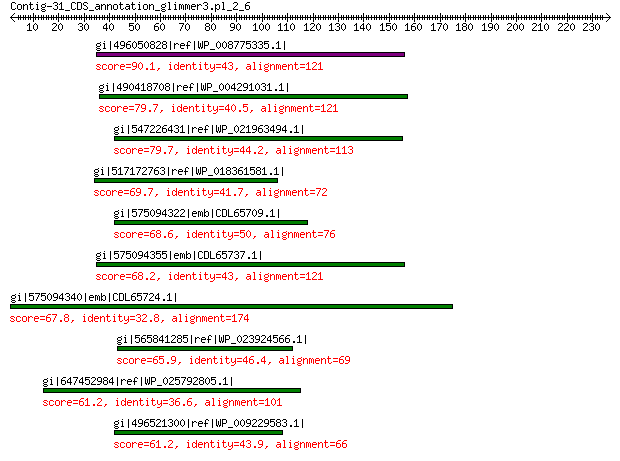
gi|496050828|ref|WP_008775335.1| hypothetical protein 90.1 3e-17
gi|490418708|ref|WP_004291031.1| hypothetical protein 79.7 6e-14
gi|547226431|ref|WP_021963494.1| predicted protein 79.7 8e-14
gi|517172763|ref|WP_018361581.1| hypothetical protein 69.7 2e-10
gi|575094322|emb|CDL65709.1| unnamed protein product 68.6 3e-10
gi|575094355|emb|CDL65737.1| unnamed protein product 68.2 5e-10
gi|575094340|emb|CDL65724.1| unnamed protein product 67.8 6e-10
gi|565841285|ref|WP_023924566.1| hypothetical protein 65.9 3e-09
gi|647452984|ref|WP_025792805.1| hypothetical protein 61.2 9e-08
gi|496521300|ref|WP_009229583.1| hypothetical protein 61.2 1e-07
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 90.1 bits (222), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 52/121 (43%), Positives = 76/121 (63%), Gaps = 2/121 (2%)
Query 35 QIPVIRNRDFELFMKRLRSNLKQQGLPHEEIRYYCVSEYGPQTLRPHWHLLLFFDSPQLT 94
+P +R D +LF+KRLR + +Q P E++RY+ V EYGP RPH+HLLLF S +
Sbjct 116 DVPYLRKTDLQLFLKRLRYYVTKQK-PSEKVRYFAVGEYGPVHFRPHYHLLLFLQSDEAL 174
Query 95 QAIRENVCKAWSYGNCDVSLSRGaaasyvasyvnsvasLPYLYTGHKEIRPRCFHSKGFG 154
Q EN+ KAW++G D +S+G ++YVASYVNS ++P ++ + P HS+ G
Sbjct 175 QICSENISKAWTFGRVDCQVSKGQCSNYVASYVNSSCTIPKVFKA-SSVCPFSVHSQKLG 233
Query 155 Q 155
Q
Sbjct 234 Q 234
>gi|490418708|ref|WP_004291031.1| hypothetical protein [Bacteroides eggerthii]
gi|217986635|gb|EEC52969.1| hypothetical protein BACEGG_02720 [Bacteroides eggerthii DSM
20697]
Length=422
Score = 79.7 bits (195), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 49/121 (40%), Positives = 71/121 (59%), Gaps = 2/121 (2%)
Query 36 IPVIRNRDFELFMKRLRSNLKQQGLPHEEIRYYCVSEYGPQTLRPHWHLLLFFDSPQLTQ 95
+P +R D +LF KR R + ++ P E++RY+ + EYGP RPH+H+LLF S + Q
Sbjct 41 LPYLRKFDLQLFFKRFRYYVAKR-FPKEKVRYFAIGEYGPVHFRPHYHILLFLQSDEALQ 99
Query 96 AIRENVCKAWSYGNCDVSLSRGaaasyvasyvnsvasLPYLYTGHKEIRPRCFHSKGFGQ 155
+ V +AW +G D LS+G +SYVA YVNS +P + T + P C HS+ GQ
Sbjct 100 VCSKVVSEAWPFGRVDCQLSKGKCSSYVAGYVNSSVLVPKVLT-LPTLCPFCVHSQKLGQ 158
Query 156 N 156
Sbjct 159 G 159
>gi|547226431|ref|WP_021963494.1| predicted protein [Prevotella sp. CAG:1185]
gi|524103383|emb|CCY83995.1| predicted protein [Prevotella sp. CAG:1185]
Length=498
Score = 79.7 bits (195), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 50/113 (44%), Positives = 68/113 (60%), Gaps = 3/113 (3%)
Query 42 RDFELFMKRLRSNLKQQGLPHEEIRYYCVSEYGPQTLRPHWHLLLFFDSPQLTQAIRENV 101
RD +LF+KR+R NL + E+IRYY VSEYGP+T R H+H+L F+D + + + + +
Sbjct 136 RDAQLFLKRVRKNLSK--YSDEKIRYYIVSEYGPKTFRAHYHVLFFYDEVKTQKVMSKVI 193
Query 102 CKAWSYGNCDVSLSRGaaasyvasyvnsvasLPYLYTGHKEIRPRCFHSKGFG 154
+AW +G D SLSRG SYVA YVN LP + G +P HS F
Sbjct 194 RQAWQFGRVDCSLSRGKCNSYVARYVNCNYCLP-RFLGDMSTKPFSCHSIRFA 245
>gi|517172763|ref|WP_018361581.1| hypothetical protein [Prevotella nanceiensis]
Length=598
Score = 69.7 bits (169), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/74 (41%), Positives = 46/74 (62%), Gaps = 2/74 (3%)
Query 34 LQIPVIRNRDFELFMKRLRSNLKQQGLPHEE--IRYYCVSEYGPQTLRPHWHLLLFFDSP 91
LQ + +D + F+KRLR + + +P E IRY+ SEYGP+T RPH+H +LF DSP
Sbjct 146 LQFATVSKKDIQNFLKRLRKKIDKLNIPQNEKKIRYFIASEYGPKTYRPHYHGVLFIDSP 205
Query 92 QLTQAIRENVCKAW 105
+ I+ + ++W
Sbjct 206 TVLSKIKAFIVESW 219
>gi|575094322|emb|CDL65709.1| unnamed protein product [uncultured bacterium]
Length=499
Score = 68.6 bits (166), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 38/92 (41%), Positives = 52/92 (57%), Gaps = 19/92 (21%)
Query 42 RDFELFMKRLRSNL-KQQGLPHEEIRYYCVSEYGPQTLRPHWHLLLFFDSPQLTQAIREN 100
RD +LF+KRLR ++ K G E+IR+Y + EYG ++LRPHWH LLFF+S L+QA +
Sbjct 145 RDIQLFLKRLRKHIYKYYG---EKIRFYIIGEYGTKSLRPHWHCLLFFNSSSLSQAFEDC 201
Query 101 V--------CKA-------WSYGNCDVSLSRG 117
V C W +G CD + G
Sbjct 202 VNVGTTSRPCSCPRFLRPFWQFGICDSKRTNG 233
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 68.2 bits (165), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 52/149 (35%), Positives = 71/149 (48%), Gaps = 31/149 (21%)
Query 35 QIPVIRNRDFELFMKRLRSNLKQQGLPHEEIRYYCVSEYGPQTLRPHWHLLLFFDSPQLT 94
+P +R RD +LF+KRLR NL + ++RY+ + EYGP RPH+H LLFFD + T
Sbjct 122 DVPYLRKRDLQLFIKRLRKNLSKYS--DAKVRYFAMGEYGPVHFRPHYHFLLFFDEIKFT 179
Query 95 QAI--------------RENVC--------------KAWSYGNCDVSLSRGaaasyvasy 126
+N C +W +G D S+G AA YV+SY
Sbjct 180 APSGHTLGEFPDWAWYDSQNKCSRSDILSVVEYCIRSSWKFGRVDAQYSKGDAAQYVSSY 239
Query 127 vnsvasLPYLYTGHKEIRPRCFHSKGFGQ 155
V+ SLP +Y RP HS+ GQ
Sbjct 240 VSGSGSLPKVYQV-SSARPFSLHSRFLGQ 267
>gi|575094340|emb|CDL65724.1| unnamed protein product [uncultured bacterium]
Length=486
Score = 67.8 bits (164), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 57/182 (31%), Positives = 87/182 (48%), Gaps = 14/182 (8%)
Query 1 MTDDVIKDILRRANGKYSYSLRKVVYPPASEWKLQIPVIRNRDFELFMKRLRSNLKQQGL 60
M +V ++ L G + S VV ++ ++ ++DF F+KRLR NL +
Sbjct 119 MPKEVFRNYLCNTTGIVTKSRNGVVLERDDN---KVGILYDKDFVNFVKRLRINLTRNYN 175
Query 61 PHEEIRYYCVSEYGPQTLRPHWHLLLFFDSPQLT-QAIRENVCKAWSYGNCD-----VSL 114
+I Y+ SEYGP T RPH+H + +FDS L+ + R V ++W + D V +
Sbjct 176 YEGKITYFKCSEYGPTTNRPHFHGIFWFDSRALSFDSFRSAVVESWKMCDKDKQYENVEI 235
Query 115 SRGaaasyvasyvnsvasLP-YLYTGHKEIRPRCFHSKGFG-QNKSFVNRPVFQKFEKYP 172
+R A + + P +L+ G +RP+ HSKGFG N F VF F
Sbjct 236 AREPATYVASYVNCLTSVPPLFLFKG---LRPKHSHSKGFGFANNLFSFSAVFTNFMAQR 292
Query 173 LT 174
LT
Sbjct 293 LT 294
>gi|565841285|ref|WP_023924566.1| hypothetical protein [Prevotella nigrescens]
gi|564729906|gb|ETD29850.1| hypothetical protein HMPREF1173_00032 [Prevotella nigrescens
CC14M]
Length=484
Score = 65.9 bits (159), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 32/74 (43%), Positives = 47/74 (64%), Gaps = 5/74 (7%)
Query 43 DFELFMKRLRSNL-----KQQGLPHEEIRYYCVSEYGPQTLRPHWHLLLFFDSPQLTQAI 97
D F KRLRS L K + +E+IRY+ SEYGP+TLRPH+H +++FDS ++ + I
Sbjct 119 DIVKFFKRLRSKLSYYFKKHHIITNEKIRYFVCSEYGPKTLRPHYHAIIWFDSEEVARVI 178
Query 98 RENVCKAWSYGNCD 111
+ + +WS G D
Sbjct 179 EKMLSSSWSNGFTD 192
>gi|647452984|ref|WP_025792805.1| hypothetical protein [Prevotella histicola]
Length=480
Score = 61.2 bits (147), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 37/105 (35%), Positives = 54/105 (51%), Gaps = 4/105 (4%)
Query 14 NGKY-SYSLRKVVYPPASEWKLQIPVIRNRDFELFMKRLRSNLKQQGLPHEE---IRYYC 69
NGK+ S + + + P E + +D + F KRLRS + + P IRY+
Sbjct 96 NGKFVSSDIARPIPPVGMEDTVCFAYPCKKDVQDFFKRLRSKIDYKLKPRGNEYRIRYFI 155
Query 70 VSEYGPQTLRPHWHLLLFFDSPQLTQAIRENVCKAWSYGNCDVSL 114
SEYGP T RPH+H +L++DS L + + + W GN D SL
Sbjct 156 CSEYGPNTFRPHYHAILWYDSEILHNELNVLIRETWKNGNTDFSL 200
>gi|496521300|ref|WP_009229583.1| hypothetical protein [Prevotella sp. oral taxon 317]
gi|288330571|gb|EFC69155.1| hypothetical protein HMPREF0670_00478 [Prevotella sp. oral taxon
317 str. F0108]
Length=569
Score = 61.2 bits (147), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/68 (43%), Positives = 42/68 (62%), Gaps = 2/68 (3%)
Query 42 RDFELFMKRLRSNLKQQGLPHE--EIRYYCVSEYGPQTLRPHWHLLLFFDSPQLTQAIRE 99
+D + F+KRLR N+ + E +IRYY SEYGP TLRPH+H ++FFD L I
Sbjct 136 KDIQNFLKRLRFNISKLYGKAESRKIRYYVASEYGPTTLRPHYHGIIFFDDASLLSEISS 195
Query 100 NVCKAWSY 107
+ ++W +
Sbjct 196 LIVRSWGF 203
Lambda K H a alpha
0.326 0.141 0.441 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 981586889820