bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-34_CDS_annotation_glimmer3.pl_2_2
Length=383
Score E
Sequences producing significant alignments: (Bits) Value
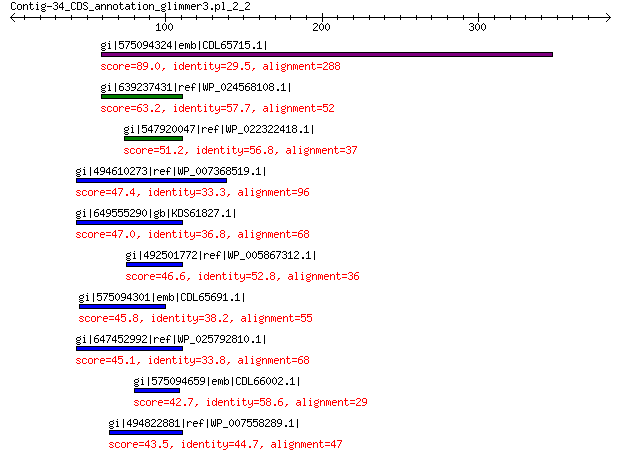
gi|575094324|emb|CDL65715.1| unnamed protein product 89.0 2e-16
gi|639237431|ref|WP_024568108.1| hypothetical protein 63.2 6e-08
gi|547920047|ref|WP_022322418.1| putative uncharacterized protein 51.2 4e-04
gi|494610273|ref|WP_007368519.1| hypothetical protein 47.4 0.011
gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 47.0 0.012
gi|492501772|ref|WP_005867312.1| hypothetical protein 46.6 0.014
gi|575094301|emb|CDL65691.1| unnamed protein product 45.8 0.043
gi|647452992|ref|WP_025792810.1| hypothetical protein 45.1 0.074
gi|575094659|emb|CDL66002.1| unnamed protein product 42.7 0.16
gi|494822881|ref|WP_007558289.1| hypothetical protein 43.5 0.20
>gi|575094324|emb|CDL65715.1| unnamed protein product [uncultured bacterium]
Length=370
Score = 89.0 bits (219), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 85/317 (27%), Positives = 156/317 (49%), Gaps = 41/317 (13%)
Query 59 KFAREERLAQQQWIEQMYE-----------KNNSYNSPAAQMQRLKEAGLNPDLMYSRGD 107
K REE + W ++M E +YN+P+A M+RLK+AGLNPDLMY G
Sbjct 32 KAQREENEKARNWQQKMAEWQVGIERENLADERAYNNPSAVMKRLKDAGLNPDLMYGSGA 91
Query 108 VGNatapeapqqaatpRYNVIPANTYGQTA----------QIAADAGLKAAQARLAsses 157
G ++ + NV PA+ G Q AA A A+ + +++
Sbjct 92 SG---LVDSNVAGSASVGNVPPADVAGPIMGTPTMMESLFQGAAYAK-TVAETKNIKADT 147
Query 158 kktdteesLLTADYLLRKARTESDIELNSSTIYVNHELGQLNHAEAEVAAKKLQEIDVAM 217
K + E + L D ++ A +++ I+L+ + QL A+AE +++ ++ +
Sbjct 148 SKKEGEVTSLNIDNFVKAASSDNAIKLSGVEV-------QLTKAQAEYTSEQKTKLISEI 200
Query 218 SEARERISTLKAQQSQ-------IDENLVQMKFDRFLRSKEFELLCKKTYQDMKESNSRL 270
++ E ++ LKAQ S+ +D + V + L ++ F+L C++ + ++E+++++
Sbjct 201 NDINEHVNLLKAQISETWARTSNLDSSTVLNRTTAILNNRRFDLECEEFARRVRETDAKV 260
Query 271 TLNAAEVQDMMATQLARVMNLNASTYM-QKKQGLLASEQTMTELYKQTGIDISNQHAKFN 329
L+ AE + ++ T A+V N++A T + Q L +++T E Y + IDI A F
Sbjct 261 NLSEAEAKSILVTMYAKVNNIDADTALKQANIRLTDAQKTQVEHYTNS-IDIHRDAAVFK 319
Query 330 FDQAKNWDSTERFTNVA 346
Q + +D +R +VA
Sbjct 320 LQQDQKYDDAQRIVSVA 336
>gi|639237431|ref|WP_024568108.1| hypothetical protein [Elizabethkingia anophelis]
Length=287
Score = 63.2 bits (152), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 30/52 (58%), Positives = 36/52 (69%), Gaps = 4/52 (8%)
Query 59 KFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLMYSRGDVGN 110
K ARE R + M+ +NN YN+P AQMQRLKEAGLNP+LMY +G GN
Sbjct 25 KIARENRA----FALDMWNRNNEYNTPLAQMQRLKEAGLNPNLMYGQGTTGN 72
>gi|547920047|ref|WP_022322418.1| putative uncharacterized protein [Parabacteroides merdae CAG:48]
gi|524592959|emb|CDD13571.1| putative uncharacterized protein [Parabacteroides merdae CAG:48]
Length=259
Score = 51.2 bits (121), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/37 (57%), Positives = 27/37 (73%), Gaps = 0/37 (0%)
Query 74 QMYEKNNSYNSPAAQMQRLKEAGLNPDLMYSRGDVGN 110
+M+ N YNSP AQM RL++AGLNP+L+Y G GN
Sbjct 24 EMWNMQNQYNSPTAQMSRLRQAGLNPNLVYGSGVTGN 60
>gi|494610273|ref|WP_007368519.1| hypothetical protein [Prevotella multiformis]
gi|324988545|gb|EGC20508.1| hypothetical protein HMPREF9141_0987 [Prevotella multiformis
DSM 16608]
Length=437
Score = 47.4 bits (111), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 32/97 (33%), Positives = 51/97 (53%), Gaps = 5/97 (5%)
Query 43 NKSNRKLSKEAFERESKFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLM 102
N++N ++++EA + + + +E Q + E+M+ + N YNSPAAQMQR +AG+NP +
Sbjct 97 NETNLQIAREANQNQYQMFQE----QNAFNERMWNQMNQYNSPAAQMQRYTDAGINPYIA 152
Query 103 YSRGDVGNatapeapqqaatpRY-NVIPANTYGQTAQ 138
GNA + A V+PA G Q
Sbjct 153 AGNVQTGNAQSALQSAPAPQQHVAQVMPATGMGDAVQ 189
>gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649557306|gb|KDS63785.1| hypothetical protein M095_3404 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649559158|gb|KDS65545.1| hypothetical protein M096_4689 [Parabacteroides distasonis str.
3999B T(B) 6]
gi|649560567|gb|KDS66875.1| hypothetical protein M095_2448 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561016|gb|KDS67303.1| hypothetical protein M095_2410 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649562727|gb|KDS68911.1| hypothetical protein M096_3341 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=288
Score = 47.0 bits (110), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 25/68 (37%), Positives = 41/68 (60%), Gaps = 4/68 (6%)
Query 43 NKSNRKLSKEAFERESKFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLM 102
NK+N +++K ++++ ++E Q +M+ N YNSP QM R++ AGLNP+L+
Sbjct 32 NKANMEIAK----YQAQWQQQENEKAYQRSLKMWNLQNEYNSPTQQMARIRAAGLNPNLV 87
Query 103 YSRGDVGN 110
Y G GN
Sbjct 88 YGNGVTGN 95
>gi|492501772|ref|WP_005867312.1| hypothetical protein [Parabacteroides distasonis]
gi|409230405|gb|EKN23269.1| hypothetical protein HMPREF1059_03254 [Parabacteroides distasonis
CL09T03C24]
Length=288
Score = 46.6 bits (109), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 19/36 (53%), Positives = 24/36 (67%), Gaps = 0/36 (0%)
Query 75 MYEKNNSYNSPAAQMQRLKEAGLNPDLMYSRGDVGN 110
M+ N YNSP QM R++ AGLNP+L+Y G GN
Sbjct 60 MWNLQNEYNSPTQQMARIRAAGLNPNLVYGNGVTGN 95
>gi|575094301|emb|CDL65691.1| unnamed protein product [uncultured bacterium]
Length=437
Score = 45.8 bits (107), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 32/55 (58%), Gaps = 0/55 (0%)
Query 45 SNRKLSKEAFERESKFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNP 99
S +K + E F +E Q+ + QM++ N YN+P AQ QRL++AG+NP
Sbjct 39 SQQKFERHQAEDARNFTHQENALQRDFARQMWKDTNDYNTPIAQKQRLEQAGMNP 93
>gi|647452992|ref|WP_025792810.1| hypothetical protein [Prevotella histicola]
Length=424
Score = 45.1 bits (105), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 23/68 (34%), Positives = 40/68 (59%), Gaps = 4/68 (6%)
Query 43 NKSNRKLSKEAFERESKFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLM 102
N++N ++++E + + +E + E+MY + N+YN+P+AQMQR EAG+NP +
Sbjct 30 NQTNLQIARETNQMNYQLFQESN----AFNEKMYHEANAYNTPSAQMQRYAEAGINPYIA 85
Query 103 YSRGDVGN 110
GN
Sbjct 86 AGNVQTGN 93
>gi|575094659|emb|CDL66002.1| unnamed protein product [uncultured bacterium]
Length=204
Score = 42.7 bits (99), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 17/29 (59%), Positives = 22/29 (76%), Gaps = 0/29 (0%)
Query 80 NSYNSPAAQMQRLKEAGLNPDLMYSRGDV 108
N YN+P QM+RL+ AGLNP+L+Y G V
Sbjct 39 NDYNNPINQMKRLQAAGLNPNLVYGSGSV 67
>gi|494822881|ref|WP_007558289.1| hypothetical protein [Bacteroides plebeius]
gi|198272097|gb|EDY96366.1| hypothetical protein BACPLE_00802 [Bacteroides plebeius DSM 17135]
Length=344
Score = 43.5 bits (101), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 21/47 (45%), Positives = 30/47 (64%), Gaps = 2/47 (4%)
Query 64 ERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLMYSRGDVGN 110
ER +Q+W M+ N YNS ++Q +RL+EAGLNP +M G G+
Sbjct 63 ERQIEQEW--DMWNAENEYNSASSQRKRLEEAGLNPYMMMDGGSAGS 107
Lambda K H a alpha
0.313 0.124 0.337 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2379307285017