bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
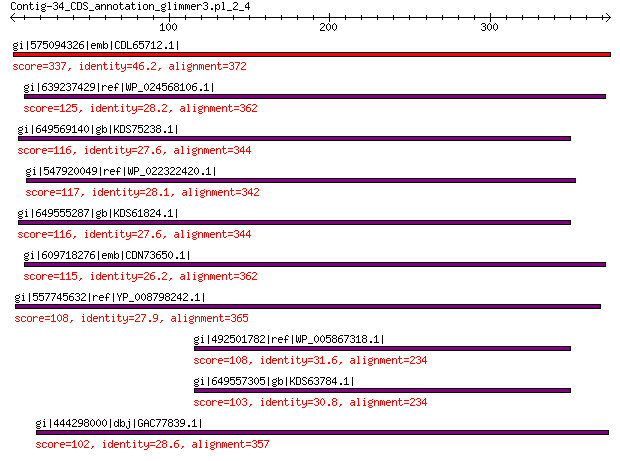
Query= Contig-34_CDS_annotation_glimmer3.pl_2_4
Length=374
Score E
Sequences producing significant alignments: (Bits) Value
gi|575094326|emb|CDL65712.1| unnamed protein product 337 4e-105
gi|639237429|ref|WP_024568106.1| hypothetical protein 125 2e-28
gi|649569140|gb|KDS75238.1| capsid family protein 116 7e-26
gi|547920049|ref|WP_022322420.1| capsid protein VP1 117 1e-25
gi|649555287|gb|KDS61824.1| capsid family protein 116 3e-25
gi|609718276|emb|CDN73650.1| conserved hypothetical protein 115 4e-25
gi|557745632|ref|YP_008798242.1| major capsid protein 108 1e-22
gi|492501782|ref|WP_005867318.1| hypothetical protein 108 1e-22
gi|649557305|gb|KDS63784.1| capsid family protein 103 2e-22
gi|444298000|dbj|GAC77839.1| major capsid protein 102 1e-20
>gi|575094326|emb|CDL65712.1| unnamed protein product [uncultured bacterium]
Length=758
Score = 337 bits (865), Expect = 4e-105, Method: Compositional matrix adjust.
Identities = 172/373 (46%), Positives = 247/373 (66%), Gaps = 16/373 (4%)
Query 3 GGADN-TTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTV 61
GG+D T L NW+ D TTA+ PQQG APLVGLT ++ + D G T
Sbjct 401 GGSDTLTPRDLRFANWQSDAYTTALTAPQQGV-APLVGLTTYEIRSVNDAGHEVTTVNTA 459
Query 62 LVDEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIET 121
+VDE+G+ Y + ++ +GE+L GV+Y P+ V + +L + SG +I
Sbjct 460 IVDEEGNAYKVDFE--SNGEALKGVNYTPLKAGEAV-------NMQSLVSPVTSGISIND 510
Query 122 LRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTV 181
R VNAYQ++LELN +GFSYK+I++GR+D+++R+D L MPE++GGI+R++ + + QTV
Sbjct 511 FRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTV 570
Query 182 DQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSK 241
+ T+ G Y +LGS+SG+A +G+T +I VFCDEES ++G++ V P+P+Y +L K
Sbjct 571 E---TTGSGSYVGSLGSQSGLATCFGNTDGSISVFCDEESIVMGIMYVMPMPVYDSLLPK 627
Query 242 DFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYD 301
Y LD + PEFD IG+QPI KE+ P+ V D ++ N FGYQRPWYEYVAK D
Sbjct 628 WLTYRERLDSFNPEFDHIGYQPIYAKELGPMQC-VQDDID-PNTVFGYQRPWYEYVAKPD 685
Query 302 SAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATA 361
AHGLF ++++NF+M R F +P+LGQ F ++ P +VN VFSVTE +DKI G + F+ TA
Sbjct 686 RAHGLFLSSLRNFIMFRSFDNVPELGQSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTA 745
Query 362 RLPISRVAIPRLD 374
+LPISRV +PRL+
Sbjct 746 QLPISRVVVPRLE 758
>gi|639237429|ref|WP_024568106.1| hypothetical protein [Elizabethkingia anophelis]
Length=546
Score = 125 bits (313), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 102/372 (27%), Positives = 163/372 (44%), Gaps = 34/372 (9%)
Query 10 YSLHQCNWERDFLTTAVPNPQQGA--------NAPLVGLTVGDVVTRADDGTYSIQKQTV 61
+ + + W D+ T+A+P Q+GA A L G+ + DG+ S T
Sbjct 195 FDIKKRAWHHDYFTSALPFAQKGAAVKMPLQMTADLFYNPGGNTFVKKPDGSLS---HTG 251
Query 62 LVDEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIET 121
EDGS V DG + V+ PV NS L T GS TI
Sbjct 252 FRLEDGS-------VPADGIGHLMVETSSTGNSNPVNIDNSSNLGVDLKTASGS--TIND 302
Query 122 LRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTV 181
LR Q++LE N R G Y + + + + L PEF+GG + + V Q
Sbjct 303 LRRAFKLQEWLEKNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKTPILISEVLQQS 362
Query 182 DQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSK 241
ST+ QG A G+ F +E Y+IGL++V P Y+Q + +
Sbjct 363 STDSTTPQGNMAGH--------GISVGKEGGFSKFFEEHGYVIGLMSVIPKTSYSQGIPR 414
Query 242 DFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYD 301
F D++ P+F+ IG QP+ KE+ N+G D+ FGY + EY
Sbjct 415 HFSKFDKFDYFWPQFEHIGEQPVYNKEIFAKNVGDYDS----GGVFGYVPRYSEYKYSPS 470
Query 302 SAHGLFRTNMKNFVMSRVF--SGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNA 359
+ HG F+ + + + R+F S P+L + F+ V+ ++++F+V + +DK + ++
Sbjct 471 TIHGDFKDTLYFWHLGRIFDSSAPPKLNRDFIEVNKSGLSRIFAVEDNSDKFYCHLYQKI 530
Query 360 TARLPISRVAIP 371
TA+ +S P
Sbjct 531 TAKRKMSYFGDP 542
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 116 bits (291), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 95/347 (27%), Positives = 154/347 (44%), Gaps = 30/347 (9%)
Query 6 DNTTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLVDE 65
+++ + LH+ WE+D+ T+A+P Q+G + G++ +G ++ QK T D
Sbjct 46 NSSLWQLHRRAWEKDYFTSALPWVQRGPEVTVPINGGGEIPVEMKEG-FAAQKITTFPDR 104
Query 66 DGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAA---LTTTEGSGFTIETL 122
K E L V + +I A + + T+ G I +
Sbjct 105 ---------KPISGSEVLYSA--PSVLSYGQIGSIKGQALIEPDNFVVNTDQMGVNINDI 153
Query 123 RYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVD 182
R NA Q++ E N R G Y + + + + L P+F+GG +S+ V QT
Sbjct 154 RTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSS 213
Query 183 QQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKD 242
STS Q A G+ ++ + +E YI+G++++ P Y Q + KD
Sbjct 214 TDSTSPQANMAG--------HGISAGVNHGFTRYFEEHGYIMGIMSIRPRTGYQQGVPKD 265
Query 243 FLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDS 302
F +D Y PEF +G Q I +E L L +D N+ TFGY + EY +
Sbjct 266 FRKFDNMDFYFPEFAHLGEQEIKNEE---LYLNESDAANEG--TFGYTPRYAEYKYSQNE 320
Query 303 AHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTD 349
HG FR NM + ++R+F P L F+ +P N+VF+ E +D
Sbjct 321 VHGDFRGNMAFWHLNRIFKEKPNLNTTFVECNPS--NRVFATAETSD 365
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 117 bits (292), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 96/346 (28%), Positives = 151/346 (44%), Gaps = 23/346 (7%)
Query 11 SLHQCNWERDFLTTAVPNPQQGANAPLVGLTVG---DVVTRADDGTYSIQKQTVLVDEDG 67
SL + WE+D+ T+A+P Q+G + G DVV + + E+G
Sbjct 205 SLRRRAWEKDYFTSALPWLQRGPEVTVPVQGAGGSMDVVYERQSDSQKWVDSSGREFENG 264
Query 68 SKYGISYKVSEDGESLVGVDYDP-VSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVN 126
Y I+ + D S + V + + + P N ++ + G I LR N
Sbjct 265 HAYDITMARANDPNSALMVAVNGGTNNRAPELDPNGTLKV----NVDEMGININDLRTSN 320
Query 127 AYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQST 186
A Q++ E N R G Y + + + + L P+F+GG +S+ V QT T
Sbjct 321 ALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRPQFLGGGRMPISVSEVLQTSSTDET 380
Query 187 SSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYN 246
S Q A G+ +N + + +E YIIG++++TP Y Q + +DF
Sbjct 381 SPQANMAGH--------GISAGINNGFKHYFEEHGYIIGIMSITPRSGYQQGVPRDFTKF 432
Query 247 GLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGL 306
+D Y PEF + Q I +E L V++ N TFGY + EY AHG
Sbjct 433 DNMDFYFPEFAHLSEQEIKNQE-----LFVSEDAAYNNGTFGYTPRYAEYKYHPSEAHGD 487
Query 307 FRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIF 352
FR N+ + ++R+F P L F+ P N+VF+ +E D F
Sbjct 488 FRGNLSFWHLNRIFEDKPNLNTTFVECKPS--NRVFATSETEDDKF 531
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 116 bits (290), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 95/347 (27%), Positives = 154/347 (44%), Gaps = 30/347 (9%)
Query 6 DNTTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLVDE 65
+++ + LH+ WE+D+ T+A+P Q+G + G++ +G ++ QK T D
Sbjct 197 NSSLWQLHRRAWEKDYFTSALPWVQRGPEVTVPINGGGEIPVEMKEG-FAAQKITTFPDR 255
Query 66 DGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAA---LTTTEGSGFTIETL 122
K E L V + +I A + + T+ G I +
Sbjct 256 ---------KPISGSEVLYSA--PSVLSYGQIGSIKGQALIEPDNFVVNTDQMGVNINDI 304
Query 123 RYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVD 182
R NA Q++ E N R G Y + + + + L P+F+GG +S+ V QT
Sbjct 305 RTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSS 364
Query 183 QQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKD 242
STS Q A G+ ++ + +E YI+G++++ P Y Q + KD
Sbjct 365 TDSTSPQANMAG--------HGISAGVNHGFTRYFEEHGYIMGIMSIRPRTGYQQGVPKD 416
Query 243 FLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDS 302
F +D Y PEF +G Q I +E L L +D N+ TFGY + EY +
Sbjct 417 FRKFDNMDFYFPEFAHLGEQEIKNEE---LYLNESDAANEG--TFGYTPRYAEYKYSQNE 471
Query 303 AHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTD 349
HG FR NM + ++R+F P L F+ +P N+VF+ E +D
Sbjct 472 VHGDFRGNMAFWHLNRIFKEKPNLNTTFVECNPS--NRVFATAETSD 516
>gi|609718276|emb|CDN73650.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=537
Score = 115 bits (289), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 95/364 (26%), Positives = 162/364 (45%), Gaps = 27/364 (7%)
Query 10 YSLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLVDEDGSK 69
+ + + W D+ T+A+P Q+G + G+V TY + QT + D G+
Sbjct 195 FKMRKRAWHHDYFTSALPFAQKGNAVKIPIFPQGNVPL-----TYEMGSQTFIKDMAGNP 249
Query 70 YGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVNAYQ 129
S+ +L V P+S ++ L +E T+ LR Q
Sbjct 250 APNKDLRSDVNGNLQDVSGQPLS-------LDPSKNLKLNMASENVS-TVNDLRRAFKLQ 301
Query 130 KFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQ 189
++LE N R G Y + + + + L PEF+GG + + V Q ST+ Q
Sbjct 302 EWLEKNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKSPIMISEVLQQSATDSTTPQ 361
Query 190 GQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLL 249
G A G GI G + F +E Y+IGL++V P Y+Q + + F +
Sbjct 362 GNMA---GHGIGIGKDGGFSR-----FFEEHGYVIGLMSVIPKTSYSQGIPRHFSKSDKF 413
Query 250 DHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRT 309
D++ P+F+ IG QP+ KE+ N+ D+ FGY + EY + HG F+
Sbjct 414 DYFWPQFEHIGEQPVYNKEIFAKNIDAFDS----EAVFGYLPRYSEYKFSPSTVHGDFKD 469
Query 310 NMKNFVMSRVF--SGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISR 367
++ + + R+F P L Q F+ D + ++++F+V + TDK + ++ TA+ +S
Sbjct 470 DLYFWHLGRIFDTDKPPVLNQSFIECDKNALSRIFAVEDDTDKFYCHLYQKITAKRKMSY 529
Query 368 VAIP 371
P
Sbjct 530 FGDP 533
>gi|557745632|ref|YP_008798242.1| major capsid protein [Marine gokushovirus]
gi|530695345|gb|AGT39902.1| major capsid protein [Marine gokushovirus]
Length=538
Score = 108 bits (270), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 102/370 (28%), Positives = 165/370 (45%), Gaps = 37/370 (10%)
Query 4 GADNTTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTYSIQKQTVLV 63
G D TTY+L + D+ T+A+P PQ+GA+ + L VT A+ S Q T+
Sbjct 195 GNDTTTYALLNRGKKHDYFTSALPWPQKGADV-TIPLGTSAPVTTANS---SNQDVTIFT 250
Query 64 DEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLR 123
G+ + S + P E T A YA+L+ E + TI LR
Sbjct 251 PNIGNTHRFLNSAS--------TNVYPGDENTD-EARRLYADLS-----EATSATINQLR 296
Query 124 YVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQ 183
A QKFLE+ R G Y ++++ +++ L PE++GG S +++ V QT
Sbjct 297 LAFATQKFLEIQARGGSRYIEVIKNHFNVTSPDARLQRPEYLGGGSSPVNISPVAQTSST 356
Query 184 QSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDF 243
+T+ QG S I S + + F E + +IG+++V Y Q L++ F
Sbjct 357 DATTPQGNL-------SAIGTTVLSGHSFTKSFT-EHTIVIGMVSVRTDLTYQQGLNRMF 408
Query 244 LYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSA 303
+ D+Y P IG Q + KE+ +T TFGYQ + EY K S
Sbjct 409 SRETIYDYYWPTLSTIGEQAVKNKEIYAQGSAADET------TFGYQERYAEYRYKPSSV 462
Query 304 HGLFRTN----MKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIF-GYVKFN 358
G FR+N ++++ ++ ++ LP LG ++ V V + +V IF K
Sbjct 463 TGKFRSNATGTLESWHYAQEYASLPLLGDSWIQVTDTNVQRTLAVASEPQFIFDSLFKLR 522
Query 359 ATARLPISRV 368
T +P++ +
Sbjct 523 CTRPMPVNSI 532
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 108 bits (270), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 74/234 (32%), Positives = 111/234 (47%), Gaps = 15/234 (6%)
Query 116 GFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMR 175
G +I LR NA Q++ E N R G Y + + + + L P+F+GG +S+
Sbjct 295 GVSINDLRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVS 354
Query 176 TVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIY 235
V QT STS Q A G+ ++ + + +E YIIG++++ P Y
Sbjct 355 EVLQTSATDSTSPQANMAGH--------GISAGVNHGFKRYFEEHGYIIGIMSIRPRTGY 406
Query 236 TQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYE 295
Q + KDF +D Y PEF +G Q I +EV + T N TFGY + E
Sbjct 407 QQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEVY-----LQQTPASNNGTFGYTPRYAE 461
Query 296 YVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTD 349
Y + HG FR NM + ++R+FS P L F+ +P N+VF+ E +D
Sbjct 462 YKYSMNEVHGDFRGNMAFWHLNRIFSESPNLNTTFVECNPS--NRVFATAETSD 513
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 103 bits (258), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 72/234 (31%), Positives = 110/234 (47%), Gaps = 15/234 (6%)
Query 116 GFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMR 175
G I +R NA Q++ E N R G Y + + + + L P+F+GG +S+
Sbjct 2 GVNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVS 61
Query 176 TVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIY 235
V QT STS Q A G+ ++ + +E YI+G++++ P Y
Sbjct 62 EVLQTSSTDSTSPQANMAG--------HGISAGVNHGFTRYFEEHGYIMGIMSIRPRTGY 113
Query 236 TQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYE 295
Q + KDF +D Y PEF +G Q I +E L L +D N+ TFGY + E
Sbjct 114 QQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEE---LYLNESDAANEG--TFGYTPRYAE 168
Query 296 YVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTD 349
Y + HG FR NM + ++R+F P L F+ +P N+VF+ E +D
Sbjct 169 YKYSQNEVHGDFRGNMAFWHLNRIFKEKPNLNTTFVECNPS--NRVFATAETSD 220
>gi|444298000|dbj|GAC77839.1| major capsid protein [uncultured marine virus]
Length=480
Score = 102 bits (253), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 102/367 (28%), Positives = 160/367 (44%), Gaps = 40/367 (11%)
Query 17 WERDFLTTAVPNPQQGANAPLVGLTVGDVVTRADDGTY-SIQKQTVLVDEDGS---KYGI 72
W++D+ T+A P Q+G P V L +GD GT S Q + V+E G +YG
Sbjct 143 WQKDYFTSARPWTQKG---PDVTLPLGDRAPIYGIGTTGSPATQNINVNETGGVNREYGA 199
Query 73 SYKVSEDGESLVGVDYDP-VSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVNAYQKF 131
++ + D DP P YA+L A T G TI +R A Q++
Sbjct 200 AWSSETTNAIVAEHDPDPGAGSDDP----GIYADLQAAT-----GGTINDIRRAFAIQRY 250
Query 132 LELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQ 191
E R G Y + ++ ++ + L PE++GG + +++ V QT + Q
Sbjct 251 QEARSRYGSRYTEYLR-YLGVNPKDARLQRPEYMGGGTTQINFSEVLQTSPEIPGEDQV- 308
Query 192 YAEALGSKSGIAGVYGS-----TSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYN 246
S+ G+ +YG SN + +E YII +L+V P +YT + + +L
Sbjct 309 ------SQFGVGDMYGHGIAAMRSNKYRRYIEEHGYIISMLSVRPKTMYTNGIHRSWLRL 362
Query 247 GLLDHYQPEFDRIGFQPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGL 306
D+YQ E + IG Q I E+ AD +TFGY + EY
Sbjct 363 TKEDYYQKELEHIGQQEIMNNEI------YADE-GAGTETFGYNDRYSEYRETPSHVSAE 415
Query 307 FRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPIS 366
FR + + M+R F P L Q F VD D ++ + + D ++ ++ AR +S
Sbjct 416 FRGILNYWHMAREFEAPPVLNQSF--VDCDATKRIHN-EQTQDALWIMIQHKMVARRLLS 472
Query 367 RVAIPRL 373
R A PR+
Sbjct 473 RNAAPRI 479
Lambda K H a alpha
0.317 0.135 0.394 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2308641853950