bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-36_CDS_annotation_glimmer3.pl_2_3
Length=318
Score E
Sequences producing significant alignments: (Bits) Value
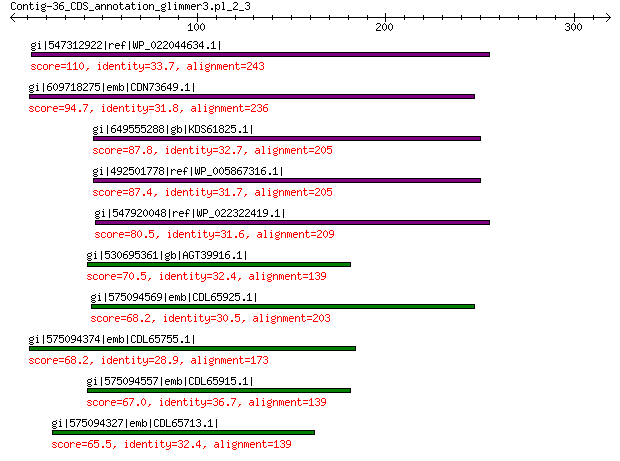
gi|547312922|ref|WP_022044634.1| putative replication initiation... 110 3e-24
gi|609718275|emb|CDN73649.1| conserved hypothetical protein 94.7 2e-19
gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 87.8 7e-17
gi|492501778|ref|WP_005867316.1| hypothetical protein 87.4 9e-17
gi|547920048|ref|WP_022322419.1| putative replication protein 80.5 2e-14
gi|530695361|gb|AGT39916.1| replication initiator 70.5 1e-10
gi|575094569|emb|CDL65925.1| unnamed protein product 68.2 1e-09
gi|575094374|emb|CDL65755.1| unnamed protein product 68.2 2e-09
gi|575094557|emb|CDL65915.1| unnamed protein product 67.0 3e-09
gi|575094327|emb|CDL65713.1| unnamed protein product 65.5 1e-08
>gi|547312922|ref|WP_022044634.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
gi|524208442|emb|CCZ76638.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
Length=320
Score = 110 bits (274), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 82/266 (31%), Positives = 132/266 (50%), Gaps = 36/266 (14%)
Query 12 CLYPKLIRNKRYLPNKKNGGNVPY------CDDKRKKAVAIGCGKCIECRKKRANEWRVR 65
C PK+I N+RY N N V Y C + + CG C C+K N++R+R
Sbjct 3 CEQPKVIVNRRYA-NMTNTEIVNYAKVYYGCFWPPDYILEVPCGYCHSCQKSYNNQYRIR 61
Query 66 LMIELKDHPENA-HFVTMTYSDESLSKFEQEEALSVASRSIELFRKRWYKRYKCGIKHFL 124
L+ EL+ +P FVT+T++D+SL KF ++ ++++ LF R+ K Y I+H+
Sbjct 62 LLYELRKYPPGTCLFVTLTFNDDSLEKFSKD-----TNKAVRLFLDRFRKVYGKQIRHWF 116
Query 125 ICELGGNDSQRMHLHGILWTEKSK-------------EEVEEVWGYGFVDYGEFVNEKTV 171
+CE G R H HGIL+ + W YGFV G +V+++T
Sbjct 117 VCEFGTLHG-RPHYHGILFNVPQALIDGYDSDMPGHHPLLASCWKYGFVFVG-YVSDETC 174
Query 172 NYIVKYIFKV---DEKHPDFVTKVWTSKGIGREYAESEGKCFNKFKGISTKDYYRMPSGL 228
+YI KY+ K D+ P +V +S GIG Y +E +K G + + +G
Sbjct 175 SYITKYVTKSINGDKVRP----RVISSFGIGSNYLNTEESSLHKL-GNQRYQPFMVLNGF 229
Query 229 KVALPIYIRNKIFNENQREELWMQKL 254
+ A+P Y NKIF++ ++ + + +L
Sbjct 230 QQAMPRYYYNKIFSDVDKQNMVVDRL 255
>gi|609718275|emb|CDN73649.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=265
Score = 94.7 bits (234), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 75/243 (31%), Positives = 110/243 (45%), Gaps = 27/243 (11%)
Query 11 MCLYPKLIRNKRYLPNKKNGGNVPYCDDKRKKAVAIGCGKCIECRKKRANEWRVRLMIEL 70
MCL P ++ N + P CGKC+ECRK R N W RL EL
Sbjct 1 MCLTPVTLKKTGATTNGYATQSFP-------------CGKCLECRKARTNSWFARLTEEL 47
Query 71 KDHPENAHFVTMTYSDESLSKFEQEEALSVASRSIELFRKRWYKRYKCGIKHFLICELGG 130
K ++AHFVT+TYSD L + +S+ R +LF KR K K IK+FL+ E G
Sbjct 48 KV-SKSAHFVTLTYSDVYLP-YSDNGLISLDYRDFQLFMKRARKLQKSKIKYFLVGEYGA 105
Query 131 NDSQRMHLHGILWTEKSKEEVEEVWGYGFVDYGEFVNEKTVNYIVKYIFK-------VDE 183
+ R H H I++ ++ + W G V G V K++ Y +KY K D
Sbjct 106 Q-TYRPHYHAIVFGVENIDAFLGEWRMGNVHAGT-VTAKSIYYTLKYCTKSITEGPDKDP 163
Query 184 KHPDFVTKVWTSKGIGREYAESEGKCFNKFKGISTKDYYRMPSGLKVALPIYIRNKIFNE 243
K SKG+G + + K+ + + G +ALP Y R+K+F++
Sbjct 164 DDDRKPEKALMSKGLGLSHLT---ESMIKYYKDDVSRSFSLLGGTTIALPRYYRDKVFSD 220
Query 244 NQR 246
++
Sbjct 221 IEK 223
>gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649560564|gb|KDS66872.1| hypothetical protein M095_2449 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561011|gb|KDS67298.1| hypothetical protein M095_2409 [Parabacteroides distasonis str.
3999B T(B) 4]
Length=284
Score = 87.8 bits (216), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 67/226 (30%), Positives = 111/226 (49%), Gaps = 28/226 (12%)
Query 45 AIGCGKCIECRKKRANEWRVRLMIELKDHPENAHFVTMTYSDESLSKFEQEEAL------ 98
A+ CG+C+ CRK + W RL E ++P + FVT+TY DE + E L
Sbjct 14 AVPCGRCVNCRKNKRQSWVYRLQAEADEYPFSL-FVTLTYDDEHIPTAMIGEDLFKTTVG 72
Query 99 SVASRSIELFRKRWYKRY-KCGIKHFLICELGGNDSQRMHLHGILW-----TEKSKEEVE 152
V+ R I+LF KR K+Y + +++FL E G R H H IL+ + + +
Sbjct 73 VVSKRDIQLFMKRLRKKYAQYRLRYFLTSEYGSQGG-RPHYHMILFGFPFTGKHGGDLLA 131
Query 153 EVWGYGFVDYGEFVNEKTVNYIVKYIFKVDEKHPDFVTKV-------WTSK--GIGREYA 203
E W GFV + K ++Y+ KY+++ PD + V SK GIG +
Sbjct 132 ECWKNGFVQ-AHPLTTKEISYVTKYMYE-KSMIPDILKGVKEYQPFMLCSKMPGIGYHFL 189
Query 204 ESEGKCFNKFKGISTKDYYRMPSGLKVALPIYIRNKIFNENQREEL 249
+ F + +DY R +G+++A+P Y +K+++++ +E L
Sbjct 190 REQ---ILDFYRLHPRDYVRAFNGMRMAMPRYYADKLYDDDMKEYL 232
>gi|492501778|ref|WP_005867316.1| hypothetical protein [Parabacteroides distasonis]
gi|409230407|gb|EKN23271.1| hypothetical protein HMPREF1059_03256 [Parabacteroides distasonis
CL09T03C24]
Length=284
Score = 87.4 bits (215), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 65/226 (29%), Positives = 109/226 (48%), Gaps = 28/226 (12%)
Query 45 AIGCGKCIECRKKRANEWRVRLMIELKDHPENAHFVTMTYSDESLSKFEQEEAL------ 98
A+ CG+C+ CRK + W RL E ++P + FVT+TY DE + E L
Sbjct 14 AVPCGRCVNCRKNKRQSWVYRLQAEADEYPFSL-FVTLTYDDEHMPTAMIGEDLFKSTVG 72
Query 99 SVASRSIELFRKRWYKRY-KCGIKHFLICELGGNDSQRMHLHGILW-----TEKSKEEVE 152
V+ R I+LF KR K+Y + +++FL E G R H H IL+ + + +
Sbjct 73 VVSKRDIQLFMKRLRKKYDQYRLRYFLTSEYGSQGG-RPHYHMILFGFPFTGKHGGDLLA 131
Query 153 EVWGYGFVDYGEFVNEKTVNYIVKYIFKVDEKHPDFVTKVWTSK---------GIGREYA 203
E W GFV + K + Y+ KY+++ PD + V + GIG +
Sbjct 132 ECWKNGFVQ-AHPLTTKEIAYVTKYMYE-KSMVPDILKDVKEYQPFMLCSRIPGIGYHFL 189
Query 204 ESEGKCFNKFKGISTKDYYRMPSGLKVALPIYIRNKIFNENQREEL 249
+ F + +DY R +G+++A+P Y +K+++++ +E L
Sbjct 190 REQ---ILDFYRLHPRDYVRAFNGMRMAMPRYYADKLYDDDMKEYL 232
>gi|547920048|ref|WP_022322419.1| putative replication protein [Parabacteroides merdae CAG:48]
gi|524592960|emb|CDD13572.1| putative replication protein [Parabacteroides merdae CAG:48]
Length=278
Score = 80.5 bits (197), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 66/236 (28%), Positives = 113/236 (48%), Gaps = 33/236 (14%)
Query 46 IGCGKCIECRKKRANEWRVRLMIELKDHPENAHFVTMTYSDESL------SKFEQEEALS 99
+ CG C+ CR+ + W RL E K++P + FVT+TY DE L S Q
Sbjct 10 VPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSL-FVTLTYDDEHLPIERIGSDLFQTNVAV 68
Query 100 VASRSIELFRKRWYKRYK-CGIKHFLICELGGNDSQRMHLHGILW-----TEKSKEEVEE 153
V+ R ++LF KR K+Y+ +++F+ E G + R H H IL+ + + + + E
Sbjct 69 VSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNG-RPHYHMILFGFPFTGKMAGDLLAE 127
Query 154 VWGYGFVDYGEFVNEKTVNYIVKYIFK--------VDEKHPDFVTKVWTSKGIGREYAES 205
W GFV + K + Y+ KY+++ DEK + GIG + ++
Sbjct 128 CWQNGFVQ-AHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPGIGFGFMKA 186
Query 206 EGKCFNKFKGISTKDYYRMPSGLKVALPIYIRNKIFNEN-------QREELWMQKL 254
+ +F +DY R +G K+A+P Y +K+++++ REE + K+
Sbjct 187 D---IIEFYRRHPRDYVRAWAGHKMAMPRYYADKLYDDDMKAFLKEMREEFFRHKM 239
>gi|530695361|gb|AGT39916.1| replication initiator [Marine gokushovirus]
Length=289
Score = 70.5 bits (171), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 45/155 (29%), Positives = 77/155 (50%), Gaps = 21/155 (14%)
Query 42 KAVAIGCGKCIECRKKRANEWRVRLMIELKDHPENAHFVTMTYSDESLSKFEQEEALSVA 101
+ + CG+CI CR + +W +R + E + H +N F+T+T+ +E ++K + E+L
Sbjct 26 RGFNLPCGQCIGCRLDYSRQWAIRCVHEAQTHEDNC-FITLTFDNEHIAKRKNPESLD-- 82
Query 102 SRSIELFRKRWYKRYKCGIKHFLICELGGNDSQRMHLHGI----------LWTEKS---- 147
+ + F KR K+Y I+ F C G+ ++R H H + LW+ K
Sbjct 83 NTEFQRFMKRLRKKYPHKIR-FFHCGEYGDQNKRPHYHALLFGHDFKDKKLWSNKGDFKL 141
Query 148 --KEEVEEVWGYGFVDYGEFVNEKTVNYIVKYIFK 180
+E+ E+W YGF G V+ T Y +Y+ K
Sbjct 142 FVSQELAELWPYGFHTIGA-VSFDTAAYCARYVMK 175
>gi|575094569|emb|CDL65925.1| unnamed protein product [uncultured bacterium]
Length=354
Score = 68.2 bits (165), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 62/248 (25%), Positives = 112/248 (45%), Gaps = 51/248 (21%)
Query 44 VAIGCGKCIECRKKRANEWRVRLMIELKDHPENAHFVTMTYSDESLSKFEQ--EEALSVA 101
+ I CGKCI CR++ A W RLM+EL+DH E+ F+T+TY D+ + + EE +S+
Sbjct 68 IEIPCGKCISCRRRYAALWTDRLMLELQDHKESC-FITLTYDDDHICCVDSPIEENVSMY 126
Query 102 SRSIELFRKRWYKRYKCGIKH--------FLICELGGNDSQRMHLHGILWTEKSKEEVE- 152
+ + + W + + ++H + C G+ + R H H IL+ + + ++
Sbjct 127 TLNKVHLQCFWKRLRQYLVRHVEPEKRIRYFACGEYGDTTFRPHYHAILFGWRPTDLIQF 186
Query 153 ----------------EVWGYGFVDYGEFVNEKTVNYIVKYIFK--------VDEK---H 185
+W G V G+ V ++ Y+ +Y K + E+
Sbjct 187 KKNFQNDTLYLSKSLASIWQNGNVMVGD-VTPESCRYVARYCLKKATGFDSEIYERLGVL 245
Query 186 PDFVTKVWTSKGIGREYAESEGKCFNKFKGISTKDYYRMPSGLKVALPIY-------IRN 238
P+FVT + GI R+Y + K+K I+ + G+ + +P Y I +
Sbjct 246 PEFVT-MSRKPGIARKYFDDHYDEIIKYKTINLS---TLKGGMSMQIPPYFIRLIEDIDS 301
Query 239 KIFNENQR 246
++F E +R
Sbjct 302 ELFKEIKR 309
>gi|575094374|emb|CDL65755.1| unnamed protein product [uncultured bacterium]
Length=487
Score = 68.2 bits (165), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 50/191 (26%), Positives = 81/191 (42%), Gaps = 31/191 (16%)
Query 11 MCLYPKLIRNKRYLPNKKNGGNVPYCDDKRKKA-VAIGCGKCIECRKKRANEWRVRLMIE 69
MC P +IRN N Y A + CG C +C+ + +W+VR E
Sbjct 1 MCFSPIIIRN-----------NSSYIHTHYTYADYVVPCGHCYDCKSAKTTDWQVRCSEE 49
Query 70 LKDHPENAHFVTMTYSDESLSKF----EQEEALSVASRSIELFRKRWYK---RYKCGIKH 122
L ++ + ++F T+T + + + R I+LF KR K +Y +K+
Sbjct 50 LNNNSQ-SYFYTLTLDPRFIDTYGTLPDGSPRYVFNKRHIQLFLKRLRKALSKYNISLKY 108
Query 123 FLICELGGNDSQRMHLHGILWTEKSKEE------VEEVWGYGFVDYGE----FVNEKTVN 172
++ EL G + R H H I + S V W GF+ G+ +N V+
Sbjct 109 VIVGEL-GETTHRPHYHAIFYLSSSVNPFKFRIMVRNSWSLGFIKSGDNNGIILNNDAVS 167
Query 173 YIVKYIFKVDE 183
Y++KY+ K D
Sbjct 168 YVIKYMHKTDS 178
>gi|575094557|emb|CDL65915.1| unnamed protein product [uncultured bacterium]
Length=354
Score = 67.0 bits (162), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 51/163 (31%), Positives = 76/163 (47%), Gaps = 27/163 (17%)
Query 42 KAVAIGCGKCIECRKKRANEWRVRLMIELKDHPENAHFVTMTYSDESLSKFEQEEALSVA 101
+ V GCGKC+ CR K EW RL++E+ H A FVT+TYS++ + E +++
Sbjct 26 RGVPFGCGKCLACRVKTRREWTSRLILEMLGHDSGA-FVTLTYSEDYVPVTESGHR-TLS 83
Query 102 SRSIELFRKRW------YKRYKCGIKHFLICELGGNDSQRMHLHGIL------------- 142
R ++LF KR KR K I+++ E G +QR H H I
Sbjct 84 LRDLQLFLKRLRRNLEERKRSKHPIRYYACGEYGTRGTQRPHYHIIFFGVSDLDLDFIKS 143
Query 143 ----WTEKSK-EEVEEVWGYGFVDYGEFVNEKTVNYIVKYIFK 180
W+E +K + + +G + E +N KTV Y Y K
Sbjct 144 VYAAWSEPAKYGQKGQTPQFGNITI-EPLNAKTVAYTAGYNMK 185
>gi|575094327|emb|CDL65713.1| unnamed protein product [uncultured bacterium]
Length=515
Score = 65.5 bits (158), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 45/152 (30%), Positives = 73/152 (48%), Gaps = 21/152 (14%)
Query 23 YLPNKKNGGNVPYCDDKRKKAVAIGCGKCIECRKKRANEWRVRLMIELKDHPENAHFVTM 82
YL N + G +P +A+ CG CI CRK++AN R ++E + + F+T+
Sbjct 63 YLLNSETGDMIPL-------YIAVPCGSCIICRKRKANALATRAIMETEITGSSPLFITL 115
Query 83 TYSDESLSKFEQEEALSVASRSIELFRKRWYKRY-KCGIKH---FLICELGGNDSQRMHL 138
TY+ E L K Q L++ ++LF KR I H +L C G++++R H
Sbjct 116 TYNPEHLPK-NQYGYLTLRKLDLQLFFKRLRSLLDNQSIPHSLRYLACGEYGSNTKRPHY 174
Query 139 HGILW---------TEKSKEEVEEVWGYGFVD 161
H +LW K++ +++ W Y VD
Sbjct 175 HLLLWGFPLSYFKDILKAQAFIQKAWSYFQVD 206
Lambda K H a alpha
0.320 0.138 0.429 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1771329643848