bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-37_CDS_annotation_glimmer3.pl_2_3
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
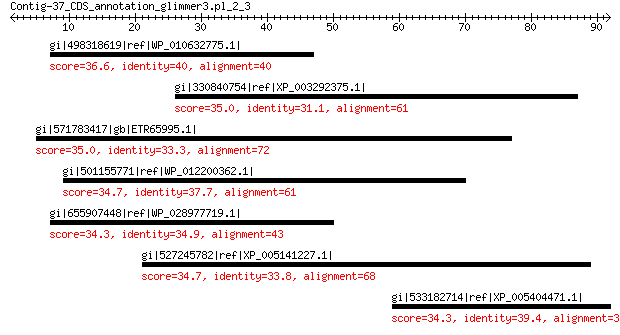
gi|498318619|ref|WP_010632775.1| hypothetical protein 36.6 1.3
gi|330840754|ref|XP_003292375.1| hypothetical protein DICPUDRAFT... 35.0 4.3
gi|571783417|gb|ETR65995.1| response regulator receiver sensor s... 35.0 4.4
gi|501155771|ref|WP_012200362.1| extracellular solute-binding pr... 34.7 5.3
gi|655907448|ref|WP_028977719.1| hypothetical protein 34.3 6.5
gi|527245782|ref|XP_005141227.1| PREDICTED: uncharacterized prot... 34.7 6.7
gi|533182714|ref|XP_005404471.1| PREDICTED: nucleolar protein 10... 34.3 9.0
>gi|498318619|ref|WP_010632775.1| hypothetical protein [Sporolactobacillus vineae]
Length=407
Score = 36.6 bits (83), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 27/40 (68%), Gaps = 4/40 (10%)
Query 7 FRVNVEKANAIYSVFKTCNVFTMLLDGTLLPVYDFSGNNV 46
F++N+EKAN++ + T N++ L+GT+L FSGN +
Sbjct 191 FKINLEKANSVIDIVNTRNLYHAFLNGTIL----FSGNKI 226
>gi|330840754|ref|XP_003292375.1| hypothetical protein DICPUDRAFT_95597 [Dictyostelium purpureum]
gi|325077382|gb|EGC31098.1| hypothetical protein DICPUDRAFT_95597 [Dictyostelium purpureum]
Length=457
Score = 35.0 bits (79), Expect = 4.3, Method: Composition-based stats.
Identities = 19/64 (30%), Positives = 32/64 (50%), Gaps = 5/64 (8%)
Query 26 VFTMLLDGTLLPVYDFSGNNVRSSLNSLRK---YYPDSVVKFHFESSFYTQNTLPCPYHF 82
+F LL G L ++ N ++ N+L+K Y PD + K+H ++ Y T+ F
Sbjct 384 IFQQLLKGLGLDSFEIIPNELKEIYNTLKKTEFYDPDEICKYHARAALYDLETI--TKQF 441
Query 83 HYPD 86
+PD
Sbjct 442 IFPD 445
>gi|571783417|gb|ETR65995.1| response regulator receiver sensor signal transduction histidine
kinase [Candidatus Magnetoglobus multicellularis str. Araruama]
Length=406
Score = 35.0 bits (79), Expect = 4.4, Method: Composition-based stats.
Identities = 24/72 (33%), Positives = 38/72 (53%), Gaps = 1/72 (1%)
Query 5 KLFRVNVEKANAIYSVFKTCNVFTMLLDGTLLPVYDFSGNNVRSSLNSLRKYYPDSVVKF 64
+L RVN E AI + K +VF+ L G++ ++ SG VRS+ N L + + + V
Sbjct 153 QLSRVNSELTTAIETREKIFSVFSDDLFGSMGSIFGLSGRIVRSAENLLNETFVKTAVNI 212
Query 65 HFESSFYTQNTL 76
S+ YTQ +
Sbjct 213 Q-RSASYTQRII 223
>gi|501155771|ref|WP_012200362.1| extracellular solute-binding protein [Lachnoclostridium phytofermentans]
gi|160880479|ref|YP_001559447.1| extracellular solute-binding protein [Lachnoclostridium phytofermentans
ISDg]
gi|160429145|gb|ABX42708.1| extracellular solute-binding protein family 1 [Lachnoclostridium
phytofermentans ISDg]
Length=431
Score = 34.7 bits (78), Expect = 5.3, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 34/62 (55%), Gaps = 6/62 (10%)
Query 9 VNVEKAN-AIYSVFKTCNVFTMLLDGTLLPVYDFSGNNVRSSLNSLRKYYPDSVVKFHFE 67
VN K N AIY++ T N F M D +LL + N+++S + K P +V F+FE
Sbjct 148 VNTVKVNDAIYAIPFTHNTFFMYYDKSLL-----NENDIKSIEGIMAKETPSNVYNFYFE 202
Query 68 SS 69
S+
Sbjct 203 SA 204
>gi|655907448|ref|WP_028977719.1| hypothetical protein [Sporolactobacillus terrae]
Length=407
Score = 34.3 bits (77), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 15/43 (35%), Positives = 27/43 (63%), Gaps = 4/43 (9%)
Query 7 FRVNVEKANAIYSVFKTCNVFTMLLDGTLLPVYDFSGNNVRSS 49
F++N+EKA ++ + T N++ L+GT+L FSGN + +
Sbjct 191 FKINLEKAKSVLDIVNTRNLYHAFLNGTIL----FSGNKITAE 229
>gi|527245782|ref|XP_005141227.1| PREDICTED: uncharacterized protein LOC101876764 [Melopsittacus
undulatus]
Length=4032
Score = 34.7 bits (78), Expect = 6.7, Method: Composition-based stats.
Identities = 23/72 (32%), Positives = 36/72 (50%), Gaps = 5/72 (7%)
Query 21 FKTCNVFTMLLDGTLLPVYDFSG-NNVRSSLNSLRKYYPDSVVKFHFESSFYTQNT---L 76
FK C+ D + +YD + ++S+L + + Y D+ + H S + NT L
Sbjct 3318 FKACHDLVKPEDFIQICIYDMCQYDGMKSALCDIVQAYVDTC-RNHGISIKWRNNTFCPL 3376
Query 77 PCPYHFHYPDCI 88
PCP H HY DC+
Sbjct 3377 PCPSHSHYTDCV 3388
>gi|533182714|ref|XP_005404471.1| PREDICTED: nucleolar protein 10 isoform X3 [Chinchilla lanigera]
Length=638
Score = 34.3 bits (77), Expect = 9.0, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 59 DSVVKFHFESSFYTQNTLPC---PYHFHYPDCILFF 91
D ++FH +S FY + +P + +HYP C L+F
Sbjct 116 DRYIEFHSQSGFYYKTRIPKFGRDFSYHYPSCDLYF 151
Lambda K H a alpha
0.327 0.140 0.443 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 437208516180