bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-3_CDS_annotation_glimmer3.pl_2_2
Length=566
Score E
Sequences producing significant alignments: (Bits) Value
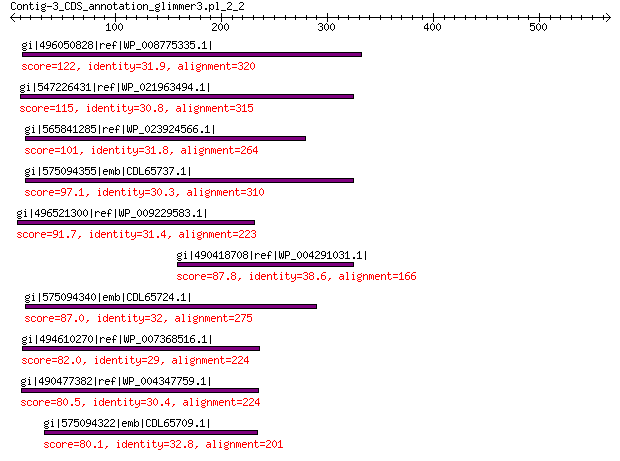
gi|496050828|ref|WP_008775335.1| hypothetical protein 122 2e-26
gi|547226431|ref|WP_021963494.1| predicted protein 115 4e-24
gi|565841285|ref|WP_023924566.1| hypothetical protein 101 2e-19
gi|575094355|emb|CDL65737.1| unnamed protein product 97.1 5e-18
gi|496521300|ref|WP_009229583.1| hypothetical protein 91.7 3e-16
gi|490418708|ref|WP_004291031.1| hypothetical protein 87.8 3e-15
gi|575094340|emb|CDL65724.1| unnamed protein product 87.0 8e-15
gi|494610270|ref|WP_007368516.1| hypothetical protein 82.0 3e-13
gi|490477382|ref|WP_004347759.1| hypothetical protein 80.5 1e-12
gi|575094322|emb|CDL65709.1| unnamed protein product 80.1 1e-12
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 122 bits (305), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 102/325 (31%), Positives = 165/325 (51%), Gaps = 49/325 (15%)
Query 12 LAECLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMNIAS-NFKYCYFVWLSYE 70
+CLHP +++N YT + + PCGHC +C K++R A ++ S K+ F+ L+Y
Sbjct 6 FCKCLHPKRIMNPYTKESMVVPCGHCQACTLAKNSR-YAFQCDLESYTAKHTLFITLTYA 64
Query 71 DQYLPYMELKSIDSLDNNRSNYFFSSVNRNLRILVPNGKDRIIEDPLFEFTHPMTSFEYQ 130
++++P F S+ R P G D +I+ E P E +
Sbjct 65 NRFIP--------------RAMFVDSIER------PYGCD-LIDKETGEILGPADLTEDE 103
Query 131 DIIVKSHGRYDFLRKCVVYPRFEDCDNRIPYCNTSDCQKFLKRLRFH-SKNKYNEEIRFY 189
R + L K ++ +PY +D Q FLKRLR++ +K K +E++R++
Sbjct 104 --------RTNLLNKFYLF-------GDVPYLRKTDLQLFLKRLRYYVTKQKPSEKVRYF 148
Query 190 GVSEYGPRTYRPHWHLLLFFNSDELTSVIQQFVSESWSYGRTTCELsrggsssyvasyvn 249
V EYGP +RPH+HLLLF SDE + + +S++W++GR C++S+G S+YVASYVN
Sbjct 149 AVGEYGPVHFRPHYHLLLFLQSDEALQICSENISKAWTFGRVDCQVSKGQCSNYVASYVN 208
Query 250 snVCLPSLYLQHKEIRARSLHSKGYGNNYVFPTQATIHEFDKMYSLLLNG---ESISFNG 306
S+ +P ++ + + S+HS+ G ++ + +K+YSL SI NG
Sbjct 209 SSCTIPKVF-KASSVCPFSVHSQKLGQGFLDCQR------EKIYSLTPENFIRSSIVLNG 261
Query 307 KAKQIYPSRSYKHTVFPRFSNLVCK 331
K K+ RS +PR V K
Sbjct 262 KYKEFDVWRSCYSFFYPRCKGFVTK 286
>gi|547226431|ref|WP_021963494.1| predicted protein [Prevotella sp. CAG:1185]
gi|524103383|emb|CCY83995.1| predicted protein [Prevotella sp. CAG:1185]
Length=498
Score = 115 bits (287), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 97/315 (31%), Positives = 157/315 (50%), Gaps = 29/315 (9%)
Query 10 YLLAECLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMNIASNFKYCYFVWLSY 69
Y L +C HP V NKYT + I CG C +CLK ++++ + L + KYC F L+Y
Sbjct 6 YPLVKCYHPRHVQNKYTGEVIQVGCGVCKACLKRRADKMSFLCAIEEQSHKYCMFATLTY 65
Query 70 EDQYLPYMELKSIDSLDNNRSNYFFSSVNRNLRILVPNGKDRIIEDPLFEFTHPMTSFEY 129
+ Y+P M + + L R ++S +R L GK ++ +++ H S +
Sbjct 66 SNDYVPRMYPEVDNELRLVR---WYSYCDR----LNEKGKLMTVD---YDYWHKCPSLDT 115
Query 130 QDIIVKSHGRYDFLRKCVVYPRFEDCDNRIPYCNTSDCQKFLKRLRFHSKNKYNEEIRFY 189
+++ + KC + D + Y + D Q FLKR+R + +E+IR+Y
Sbjct 116 YVLMLTA--------KC-------NLDGYLSYTSKRDAQLFLKRVRKNLSKYSDEKIRYY 160
Query 190 GVSEYGPRTYRPHWHLLLFFNSDELTSVIQQFVSESWSYGRTTCELsrggsssyvasyvn 249
VSEYGP+T+R H+H+L F++ + V+ + + ++W +GR C LSRG +SYVA YVN
Sbjct 161 IVSEYGPKTFRAHYHVLFFYDEVKTQKVMSKVIRQAWQFGRVDCSLSRGKCNSYVARYVN 220
Query 250 snVCLPSLYLQHKEIRARSLHSKGYGNNYVFPTQATIHEFDKMYSLLLNGESISFNGKAK 309
N CLP +L + S HS + + I++ + +GE NG
Sbjct 221 CNYCLPR-FLGDMSTKPFSCHSIRFALGIHQSQKEEIYKGSVDDFIYQSGE---INGNYV 276
Query 310 QIYPSRSYKHTVFPR 324
+ P R+ T FP+
Sbjct 277 EFMPWRNLSCTFFPK 291
>gi|565841285|ref|WP_023924566.1| hypothetical protein [Prevotella nigrescens]
gi|564729906|gb|ETD29850.1| hypothetical protein HMPREF1173_00032 [Prevotella nigrescens
CC14M]
Length=484
Score = 101 bits (251), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 84/274 (31%), Positives = 131/274 (48%), Gaps = 55/274 (20%)
Query 15 CLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMNIASNFKYCYFVWLSYEDQYL 74
C HP +++N YT++ ++ C C CL K++ + N Y FV L+Y++++L
Sbjct 9 CEHPKRIINPYTHERVWVACRRCKCCLNKKTSAWSGRVANECKLHAYSAFVTLTYDNEHL 68
Query 75 PYMELKSIDSLDNNRSNYFFSSVNRNLRILVPNGKDRIIEDPLFEFTHPMTSFEYQDIIV 134
P + + + N R ++S +R+ + + +IV
Sbjct 69 PLYQPECM----NERGEMVWTS-------------NRLCD---------------EKVIV 96
Query 135 KSHGRYDFLRKCVVYPRFEDCD-NRIPYCNTSDCQKFLKRLRFHSKNKY---------NE 184
G YDF++ + D + YC SD KF KRLR SK Y NE
Sbjct 97 ---GNYDFIK-------VSNSDVQAVAYCCKSDIVKFFKRLR--SKLSYYFKKHHIITNE 144
Query 185 EIRFYGVSEYGPRTYRPHWHLLLFFNSDELTSVIQQFVSESWSYGRTTCELsrggsssyv 244
+IR++ SEYGP+T RPH+H +++F+S+E+ VI++ +S SWS G T E + YV
Sbjct 145 KIRYFVCSEYGPKTLRPHYHAIIWFDSEEVARVIEKMLSSSWSNGFTDFEYVNSTAPQYV 204
Query 245 asyvnsnVCLPSLYLQHKEIRARSLHSKGYGNNY 278
A YV+ N LP + LQH R L S+ Y
Sbjct 205 AKYVSGNSVLPEI-LQHDACRTFHLQSQAPSVGY 237
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 97.1 bits (240), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 94/341 (28%), Positives = 150/341 (44%), Gaps = 70/341 (21%)
Query 15 CLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMNIASNFKYCYFVWLSYEDQYL 74
CL P +V N Y ND++ PCG C +C +K++R AS K+C F L+Y + Y+
Sbjct 11 CLEPKRVFNPYLNDWLLVPCGKCRACQCSKASRYKLQIQLEASQHKFCIFGTLTYANTYI 70
Query 75 PYMELKSIDSLDNNRSNYFFSSVNRNLRILVPNGKDRIIEDPLFEFTHPMTSFEYQDIIV 134
P + L N+++ F VN D+ + L P S++ + ++
Sbjct 71 PRLSLVPY----NDKT---FGVVNGYEMC------DKETGEYLGYLDSP--SYDVESLLD 115
Query 135 KSHGRYDFLRKCVVYPRFEDCDNRIPYCNTSDCQKFLKRLRFHSKNKYNEEIRFYGVSEY 194
K H F D +PY D Q F+KRLR + + ++R++ + EY
Sbjct 116 KLH-------------LFGD----VPYLRKRDLQLFIKRLRKNLSKYSDAKVRYFAMGEY 158
Query 195 GPRTYRPHWHLLLFFNSDELT----------------------------SVIQQFVSESW 226
GP +RPH+H LLFF+ + T SV++ + SW
Sbjct 159 GPVHFRPHYHFLLFFDEIKFTAPSGHTLGEFPDWAWYDSQNKCSRSDILSVVEYCIRSSW 218
Query 227 SYGRTTCELsrggsssyvasyvnsnVCLPSLYLQHKEIRARSLHSKGYGNNYVFPTQATI 286
+GR + S+G ++ YV+SYV+ + LP +Y Q R SLHS+ G ++
Sbjct 219 KFGRVDAQYSKGDAAQYVSSYVSGSGSLPKVY-QVSSARPFSLHSRFLGQGFL------A 271
Query 287 HEFDKMYSLLLNG---ESISFNGKAKQIYPSRSYKHTVFPR 324
HE +K+Y + S+ NG K RS +P+
Sbjct 272 HECEKVYETPVRDFVKRSVELNGSNKDFNLWRSCYSVFYPK 312
>gi|496521300|ref|WP_009229583.1| hypothetical protein [Prevotella sp. oral taxon 317]
gi|288330571|gb|EFC69155.1| hypothetical protein HMPREF0670_00478 [Prevotella sp. oral taxon
317 str. F0108]
Length=569
Score = 91.7 bits (226), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 70/229 (31%), Positives = 108/229 (47%), Gaps = 33/229 (14%)
Query 8 DKYLLAECLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMNIASNFKYCYFVWL 67
D + CL PV V N++T D ++ PCG C +C+ +++ + N KY L
Sbjct 4 DLKIFGNCLCPVHVHNRWTRDEMFVPCGRCEACVNAAASKQSKRVRNEIMQHKYSVMFTL 63
Query 68 SYEDQYLPYMELKSIDSLDNNRSNYFFSSVNRNLRILVPNGKDRIIEDPLFEFTHPMTSF 127
+Y ++++P E LDNN + L P G R E LF + P+ F
Sbjct 64 TYNNEFIPRWE----RFLDNN-----------DCPQLRPIG--RCAE--LFP-SCPLNYF 103
Query 128 EYQDIIVKSHGRYDFLRKCVVYPRFEDCDNRIPY--CNTSDCQKFLKRLRFHSKNKYNE- 184
+ K G++ + P+ E+ ++ + C D Q FLKRLRF+ Y +
Sbjct 104 D------KVTGKWSIDLDTFL-PKIENDEHTEVFASCCKKDIQNFLKRLRFNISKLYGKA 156
Query 185 ---EIRFYGVSEYGPRTYRPHWHLLLFFNSDELTSVIQQFVSESWSYGR 230
+IR+Y SEYGP T RPH+H ++FF+ L S I + SW + R
Sbjct 157 ESRKIRYYVASEYGPTTLRPHYHGIIFFDDASLLSEISSLIVRSWGFQR 205
>gi|490418708|ref|WP_004291031.1| hypothetical protein [Bacteroides eggerthii]
gi|217986635|gb|EEC52969.1| hypothetical protein BACEGG_02720 [Bacteroides eggerthii DSM
20697]
Length=422
Score = 87.8 bits (216), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 64/170 (38%), Positives = 95/170 (56%), Gaps = 11/170 (6%)
Query 159 IPYCNTSDCQKFLKRLRFHSKNKY-NEEIRFYGVSEYGPRTYRPHWHLLLFFNSDELTSV 217
+PY D Q F KR R++ ++ E++R++ + EYGP +RPH+H+LLF SDE V
Sbjct 41 LPYLRKFDLQLFFKRFRYYVAKRFPKEKVRYFAIGEYGPVHFRPHYHILLFLQSDEALQV 100
Query 218 IQQFVSESWSYGRTTCELsrggsssyvasyvnsnVCLPSLYLQHKEIRARSLHSKGYGNN 277
+ VSE+W +GR C+LS+G SSYVA YVNS+V +P + L + +HS+ G
Sbjct 101 CSKVVSEAWPFGRVDCQLSKGKCSSYVAGYVNSSVLVPKV-LTLPTLCPFCVHSQKLGQG 159
Query 278 YVFPTQATIHEFDKMYSLL---LNGESISFNGKAKQIYPSRSYKHTVFPR 324
++ +A K+YSL SI NG+ K+ RS FP+
Sbjct 160 FLQSERA------KVYSLTPEQFVKRSIVINGRYKEFDVWRSAYAYFFPK 203
>gi|575094340|emb|CDL65724.1| unnamed protein product [uncultured bacterium]
Length=486
Score = 87.0 bits (214), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 88/293 (30%), Positives = 136/293 (46%), Gaps = 28/293 (10%)
Query 15 CLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMN-IASNFKYCYFVWLSYEDQY 73
C + +KV NKY Y CGHC SCL+ K+N+ +N + + FV L+Y++++
Sbjct 6 CTNRIKVTNKYVGRSFYVDCGHCPSCLQRKANKSCCKIINEYGRPYSFMCFVTLTYDNEH 65
Query 74 LPYMELKSIDSLDNNRSNYFFSSVNRNLRILVPNGKDRI---------IEDPLFEFTHPM 124
+PY+ + S +Y+ R+ RI +G + + + D +F P
Sbjct 66 IPYIHPDTDYSHLYVGKSYYV----RHSRIFDKDGVENLPLGVYRNGKLIDTVFLPEMPK 121
Query 125 TSFEYQDIIVKSHGRYDFLRKCVVYPRFEDCDNRIPYCNTSDCQKFLKRLRFHSKNKYNE 184
F ++ + + G R VV E DN++ D F+KRLR + YN
Sbjct 122 EVF--RNYLCNTTGIVTKSRNGVV---LERDDNKVGILYDKDFVNFVKRLRINLTRNYNY 176
Query 185 E--IRFYGVSEYGPRTYRPHWHLLLFFNSDELT-SVIQQFVSESWSYGRTTCELsrggss 241
E I ++ SEYGP T RPH+H + +F+S L+ + V ESW + +
Sbjct 177 EGKITYFKCSEYGPTTNRPHFHGIFWFDSRALSFDSFRSAVVESWKMCDKDKQYENVEIA 236
Query 242 syvasyvnsnV-CL---PSLYLQHKEIRARSLHSKGYG-NNYVFPTQATIHEF 289
A+YV S V CL P L+L K +R + HSKG+G N +F A F
Sbjct 237 REPATYVASYVNCLTSVPPLFL-FKGLRPKHSHSKGFGFANNLFSFSAVFTNF 288
>gi|494610270|ref|WP_007368516.1| hypothetical protein [Prevotella multiformis]
gi|324988542|gb|EGC20505.1| hypothetical protein HMPREF9141_0984 [Prevotella multiformis
DSM 16608]
Length=479
Score = 82.0 bits (201), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 65/230 (28%), Positives = 109/230 (47%), Gaps = 44/230 (19%)
Query 12 LAECLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMNIASNFKYCYFVWLSYED 71
+ CL P ++ NKY ++ +Y PC C+ C + ++ + N ++ FV L+Y++
Sbjct 7 INSCLSPKRIYNKYIDETLYVPCRKCFRCRDSYASDWSRRIENECREHRFSLFVTLTYDN 66
Query 72 QYLPYMELKSIDSLDNNRSNYFFSSVNRNLRILVPNGKDRIIEDPLFEFTHPMTSFEYQD 131
+++P + +D D + +F NR L +GK + D + P + +D
Sbjct 67 EHIPLFQPLVMD--DGSHPVWF---SNR----LSESGK--FLSDSVCRSLPPQ---KMED 112
Query 132 IIVKSHGRYDFLRKCVVYPRFEDCDNRIPYCNTSDCQKFLKRLRFH-----SKNKYNE-E 185
+ C YP C D Q + KRLR +KNK NE
Sbjct 113 EV------------CFAYP-----------CK-KDVQDWFKRLRSAVDYQLNKNKSNEFR 148
Query 186 IRFYGVSEYGPRTYRPHWHLLLFFNSDELTSVIQQFVSESWSYGRTTCEL 235
IR++ SEYGPRT+RPH+H +L+++S+EL I + + E+W G + L
Sbjct 149 IRYFICSEYGPRTFRPHYHAILWYDSEELQRNIGRLIRETWKNGNSVFSL 198
>gi|490477382|ref|WP_004347759.1| hypothetical protein [Prevotella buccalis]
gi|281300711|gb|EFA93042.1| hypothetical protein HMPREF0650_1078 [Prevotella buccalis ATCC
35310]
Length=582
Score = 80.5 bits (197), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 68/236 (29%), Positives = 109/236 (46%), Gaps = 37/236 (16%)
Query 11 LLAECLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMNIASNFKYCYFVWLSYE 70
++ CL+P KV N + ++Y C C +CL K+ + A KY F L+Y+
Sbjct 9 IVGNCLNPRKVYNPSLHGWMYCSCDKCTACLNQKATTLSNRARAEIEQHKYSVFFTLTYD 68
Query 71 DQYLPYMELKSIDSLDNNRSNYFFSSVNRNLRILVPNGKDRIIEDP---LFEFTHPMTSF 127
+++LP E+ F N ++ P G R+++D + + P+ +
Sbjct 69 NEHLPKYEV--------------FQDSNEVIQYR-PIG--RLVDDSSSDMLSNSCPINKY 111
Query 128 E-YQDIIVKSHGRYDFLRKCVVYP--RFEDCDNRIPYCNTSDCQKFLKRLRFH-----SK 179
Y+++ Y F + P +ED + C D Q FLKRLR+ +
Sbjct 112 NNYENL-------YQFDESTFIPPIENYEDIYHFGVVCK-KDIQNFLKRLRWRISKIPNI 163
Query 180 NKYNEEIRFYGVSEYGPRTYRPHWHLLLFFNSDELTSVIQQFVSESWS-YGRTTCE 234
K +IR+Y SEYGP TYRPH+H +LFF+S ++ I+ + SW Y R E
Sbjct 164 TKDESKIRYYISSEYGPTTYRPHYHGILFFDSKKILDKIKSLIVMSWGKYERQQGE 219
>gi|575094322|emb|CDL65709.1| unnamed protein product [uncultured bacterium]
Length=499
Score = 80.1 bits (196), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 66/215 (31%), Positives = 96/215 (45%), Gaps = 40/215 (19%)
Query 33 PCGHCYSCLKNKSNRDTALAMNIAS-NFKYCYFVWLSYEDQYLPYMELKSIDSLDNN--- 88
PCG C +C NK + +L + + KYCYF+ L+Y+D LP + +D+
Sbjct 25 PCGKCIACHNNKRS-SLSLKLRLEEYTSKYCYFLTLTYDDDNLPLFSV-GLDTCATEFVR 82
Query 89 --------RSNYFFSSVNRNLRILVPNGKDRIIEDPLFEFTHPMTSFE--YQDIIVKSHG 138
R++ F S +L + D D + ++ + ++E Y V HG
Sbjct 83 IYPYSERLRNDSFISDFCSDL-----HNFDNDFVDKMDYYSDYVINYESKYHKSCVYGHG 137
Query 139 RYDFLRKCVVYPRFEDCDNRIPYCNTSDCQKFLKRLRFHSKNKYNEEIRFYGVSEYGPRT 198
Y L Y R D Q FLKRLR H Y E+IRFY + EYG ++
Sbjct 138 LYALL-----YYR--------------DIQLFLKRLRKHIYKYYGEKIRFYIIGEYGTKS 178
Query 199 YRPHWHLLLFFNSDELTSVIQQFVSESWSYGRTTC 233
RPHWH LLFFNS L+ + V+ + +C
Sbjct 179 LRPHWHCLLFFNSSSLSQAFEDCVNVGTTSRPCSC 213
Lambda K H a alpha
0.324 0.139 0.434 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4156673856930