bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-3_CDS_annotation_glimmer3.pl_2_7
Length=168
Score E
Sequences producing significant alignments: (Bits) Value
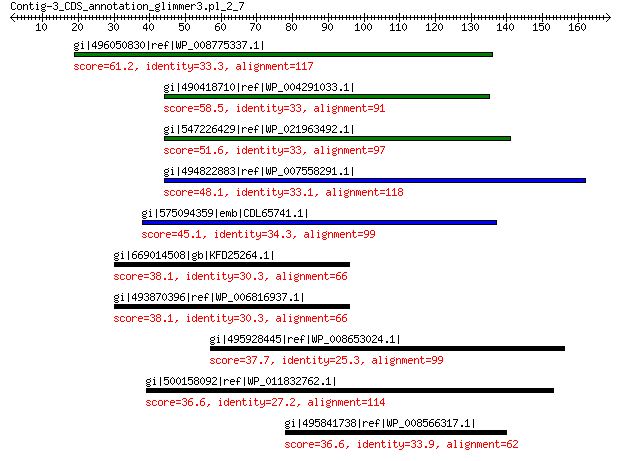
gi|496050830|ref|WP_008775337.1| hypothetical protein 61.2 5e-09
gi|490418710|ref|WP_004291033.1| hypothetical protein 58.5 4e-08
gi|547226429|ref|WP_021963492.1| putative uncharacterized protein 51.6 1e-05
gi|494822883|ref|WP_007558291.1| hypothetical protein 48.1 1e-04
gi|575094359|emb|CDL65741.1| unnamed protein product 45.1 0.002
gi|669014508|gb|KFD25264.1| putative oxidoreductase Fe-S subunit 38.1 2.4
gi|493870396|ref|WP_006816937.1| selenate reductase 38.1 2.4
gi|495928445|ref|WP_008653024.1| outer membrane efflux protein 37.7 2.9
gi|500158092|ref|WP_011832762.1| iron transporter 36.6 7.3
gi|495841738|ref|WP_008566317.1| phage tail protein 36.6 8.7
>gi|496050830|ref|WP_008775337.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448894|gb|EEO54685.1| hypothetical protein BSCG_01610 [Bacteroides sp. 2_2_4]
Length=154
Score = 61.2 bits (147), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 39/121 (32%), Positives = 66/121 (55%), Gaps = 11/121 (9%)
Query 19 CTPIREVQICPICASSEPDI--VLEELPTEPFRFEKVGTEENEAVRVRSDVSMLLHAADM 76
TP+RE I S E EE P + F F++V + + ++R+ SD+ ML + +
Sbjct 11 VTPVRE----NIVGSKEMQCSEFREESPVDQFLFQEVSVDGDTSIRLSSDIYMLFNQQRL 66
Query 77 AKKYGTGFVKSM--IEMHRPKSSGLQSEMDLMSDAQILDTIKSRHLQSPSELIAWSEYLI 134
K T ++ I + P+ + L+S++ D Q++ +KSR +QS SEL+AWS YL+
Sbjct 67 DKLSQTSLLEYFNNISVTEPRFNELRSKL---GDEQLISFVKSRFIQSKSELMAWSNYLM 123
Query 135 D 135
+
Sbjct 124 N 124
>gi|490418710|ref|WP_004291033.1| hypothetical protein [Bacteroides eggerthii]
gi|217986637|gb|EEC52971.1| hypothetical protein BACEGG_02722 [Bacteroides eggerthii DSM
20697]
Length=155
Score = 58.5 bits (140), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 55/93 (59%), Gaps = 5/93 (5%)
Query 44 PTEPFRFEKVGTEENEAVRVRSDVSMLLHAADMAKKYGTGFVKSM--IEMHRPKSSGLQS 101
P F F+++ + +++R+ SD+ ML + + K + V+ + + PK S L+
Sbjct 35 PLHEFMFQEIECDGKKSIRITSDIYMLFNQQRLDKLTRSQLVEYFDNLSVSEPKMSDLRK 94
Query 102 EMDLMSDAQILDTIKSRHLQSPSELIAWSEYLI 134
+M +D Q+ +KSR +Q+PSEL+AWS+YL+
Sbjct 95 KM---TDEQLCSFVKSRFIQTPSELMAWSQYLM 124
>gi|547226429|ref|WP_021963492.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103381|emb|CCY83992.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=152
Score = 51.6 bits (122), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 32/101 (32%), Positives = 53/101 (52%), Gaps = 5/101 (5%)
Query 44 PTEPFRFEKV----GTEENEAVRVRSDVSMLLHAADMAKKYGTGFVKSMIEMHRPKSSGL 99
P F EKV G+++ A+ D+ ML + + G ++ ++ P+S L
Sbjct 29 PIREFMTEKVTYFNGSDKKTAIAYVDDIYMLFNQNRL-DSVGRDTIQKWLDGLTPRSDSL 87
Query 100 QSEMDLMSDAQILDTIKSRHLQSPSELIAWSEYLIDQAKSI 140
+ ++D Q+++ KSR++QS SEL+AWSEYL SI
Sbjct 88 AKLRENVTDEQLMEICKSRYIQSSSELLAWSEYLNANYASI 128
>gi|494822883|ref|WP_007558291.1| hypothetical protein [Bacteroides plebeius]
gi|198272098|gb|EDY96367.1| hypothetical protein BACPLE_00803 [Bacteroides plebeius DSM 17135]
Length=140
Score = 48.1 bits (113), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 39/122 (32%), Positives = 64/122 (52%), Gaps = 6/122 (5%)
Query 44 PTEPFRFEKVGTE--ENEAVRVRSDVSMLLHAADMAKKYGTGFVKSMIEMHRPKSSGLQS 101
P E F E+V T AV R+D+ M+L+ + F + H +S L
Sbjct 12 PVEDFYREEVQTPLGSTPAVTYRNDIYMILNQRRLDSMTLAQFSDYLD--HDRSASQLSQ 69
Query 102 EMDLMSDAQILDTIKSRHLQSPSELIAWSEYL-IDQAKSIED-EASRIALERESAGTPQE 159
+ MSD Q+ +KSR++Q PSEL AW+ YL I+ +K I+ E + A++++++ T
Sbjct 70 MREKMSDEQLHQFVKSRYIQHPSELRAWASYLDIEYSKEIQKLEEAVEAVKKQTSNTDPS 129
Query 160 PN 161
P+
Sbjct 130 PD 131
>gi|575094359|emb|CDL65741.1| unnamed protein product [uncultured bacterium]
Length=156
Score = 45.1 bits (105), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 34/100 (34%), Positives = 49/100 (49%), Gaps = 2/100 (2%)
Query 38 IVLEELPTEPFRFEKVGTEEN-EAVRVRSDVSMLLHAADMAKKYGTGFVKSMIEMHRPKS 96
I E+ P E F +K ++ E+V + DV ML + + + + R S
Sbjct 25 IFREKSPVENFYIQKHKSKYGCESVSIVDDVYMLFNQQRLDRMTNAALADWLSRTSRTDS 84
Query 97 SGLQSEMDLMSDAQILDTIKSRHLQSPSELIAWSEYLIDQ 136
L S MSD Q+ IKSR++QS SEL+A+S YL Q
Sbjct 85 Q-LASLRSRMSDVQLGAFIKSRYIQSRSELLAYSSYLESQ 123
>gi|669014508|gb|KFD25264.1| putative oxidoreductase Fe-S subunit [Yokenella regensburgei
ATCC 49455]
Length=1034
Score = 38.1 bits (87), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 37/68 (54%), Gaps = 7/68 (10%)
Query 30 ICASSEPDIVLEELPTEPFRFE--KVGTEENEAVRVRSDVSMLLHAADMAKKYGTGFVKS 87
I EPD+ +E+L + F++ VGT++N +++R D + +L + ++Y G
Sbjct 619 IVYGCEPDLTVEKLQEDGFKYVLVGVGTDKNSGIKLRGDNANVLKSLPFLREYNRG---- 674
Query 88 MIEMHRPK 95
+EMH K
Sbjct 675 -VEMHLGK 681
>gi|493870396|ref|WP_006816937.1| selenate reductase [Yokenella regensburgei]
gi|364574130|gb|EHM51603.1| putative selenate reductase, YgfK subunit [Yokenella regensburgei
ATCC 43003]
Length=1042
Score = 38.1 bits (87), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 37/68 (54%), Gaps = 7/68 (10%)
Query 30 ICASSEPDIVLEELPTEPFRFE--KVGTEENEAVRVRSDVSMLLHAADMAKKYGTGFVKS 87
I EPD+ +E+L + F++ VGT++N +++R D + +L + ++Y G
Sbjct 627 IVYGCEPDLTVEKLQEDGFKYVLVGVGTDKNSGIKLRGDNANVLKSLPFLREYNRG---- 682
Query 88 MIEMHRPK 95
+EMH K
Sbjct 683 -VEMHLGK 689
>gi|495928445|ref|WP_008653024.1| outer membrane efflux protein [Rhodopirellula europaea]
gi|448888806|gb|EMB19108.1| outer membrane efflux protein [Rhodopirellula europaea 6C]
Length=629
Score = 37.7 bits (86), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 25/99 (25%), Positives = 52/99 (53%), Gaps = 9/99 (9%)
Query 57 ENEAVRVRSDVSMLLHAADMAKKYGTGFVKSMIEMHRPKSSGLQSEMDLMSDAQILDTIK 116
E+E + V + VS ++ ++ K GT + ++ +RP G S++DL +D ++
Sbjct 530 ESEQLFVTAQVSSIISLIELQKAMGTLLMTEGVQPYRP---GNTSQIDLSR----IDPVE 582
Query 117 SRHLQSPSELIAWSEYLIDQAKSIEDEASRIALERESAG 155
H+ +P+++ WS+ ++ +S+ E I E S+G
Sbjct 583 INHMHAPTQVTQWSQQVV--GESLIGEVGLIEFEDSSSG 619
>gi|500158092|ref|WP_011832762.1| iron transporter [Methanocorpusculum labreanum]
gi|124485212|ref|YP_001029828.1| hypothetical protein Mlab_0385 [Methanocorpusculum labreanum
Z]
gi|124362753|gb|ABN06561.1| small GTP-binding protein [Methanocorpusculum labreanum Z]
Length=624
Score = 36.6 bits (83), Expect = 7.3, Method: Composition-based stats.
Identities = 31/120 (26%), Positives = 58/120 (48%), Gaps = 9/120 (8%)
Query 39 VLEELPTEPFRFEKV-----GTEENEAVRV-RSDVSMLLHAADMAKKYGTGFVKSMIEMH 92
V+E + TE EK+ G E + V R D + A ++ K + +++ +
Sbjct 140 VIETIATEGKNLEKIADYVIGDELPVPIIVPRYDHHIEAAARNLQKYHNLSLAEALFALE 199
Query 93 RPKSSGLQSEMDLMSDAQILDTIKSRHLQSPSELIAWSEYLIDQAKSIEDEASRIALERE 152
S L E+ + S A I I+ RH+ SP ++I + + + AK+I DE + +A +++
Sbjct 200 NVSLSTLSPEV-MESAAAISSEIEKRHMMSPDQIIGANRH--NTAKAISDEVTTLAPQKK 256
>gi|495841738|ref|WP_008566317.1| phage tail protein [Prevotella maculosa]
gi|371949345|gb|EHO67210.1| hypothetical protein HMPREF9944_02311 [Prevotella maculosa OT
289]
Length=1545
Score = 36.6 bits (83), Expect = 8.7, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 36/65 (55%), Gaps = 4/65 (6%)
Query 78 KKYGTGFVKSMIEMHRPKSSGLQSEM---DLMSDAQILDTIKSRHLQSPSELIAWSEYLI 134
++YG F ++ I+MH + SGL M L+ D+ +LDT+K R + S E + +I
Sbjct 733 QQYGCQFARADIDMHSSRQSGLNGSMLIDSLVVDSVLLDTVKFR-IHSAEERGIYRAEVI 791
Query 135 DQAKS 139
+ K+
Sbjct 792 NNKKN 796
Lambda K H a alpha
0.312 0.127 0.359 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 438393334158